Structure and application of Enterovirus 71 3C protease
A protease, enterovirus technology, applied in the field of 3C protease structure, can solve the problem of difficulty in screening specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Obtaining of EV71 3C protease
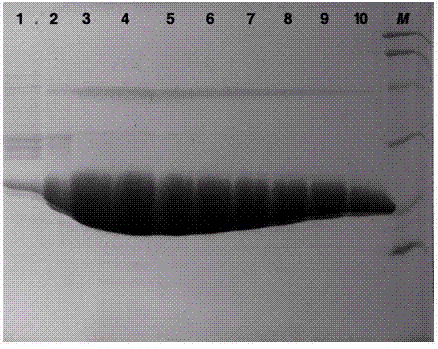
[0033] We successfully expressed the 3C protease of EV71 virus in vitro, and obtained high-purity and active enzyme protein by affinity chromatography and molecular exclusion chromatography.
[0034] The amino acid sequence of EV71 3C protease is shown in SEQ ID NO.1, or the sequence shown in SEQ ID NO.1 is derived from the amino acid sequence of SEQ ID NO.1 through substitution, deletion or addition of one or several amino acids and having 3C protease activity of protein.
[0035] Protein sequence SEQ ID NO.1:
[0036] GPSLDFALSLLRRNIRQVQTDQGHFTMLGVRDRLAVLPRHSQPGKTIWIEHKLVNVLDAVELVDEQGVNLELTLITLDTNEKFRDITKFIPENISTASDA
[0037] 1. Plasmid construction:
[0038] We artificially synthesized the DNA fragment encoding 3C protease, ligated it into the pET-21a vector with Nde1 and Xho1 restriction sites, and inserted a 6-histidine tag (hexa-His-tag) behind the encoding fragment of the enzyme protein. The coding sequence is foll...
Embodiment 2
[0050] Example 2 Structure of EV71 3C protease
[0051] 1. Protein crystallization conditions:
[0052] We used the hang-drop vapor-diffusion method and optimized the conditions to obtain crystals with good diffraction quality. The protein preparation solution used for crystallization was all protein concentration 10mg / ml. The conditions are as follows:
[0053] The crystallization conditions of EV71-3C-C147A: 0.1 M Tris-HCl (pH8.5), 20mM lithium sulfate and 25% (w / v) PEG 5000 MME; the crystal growth of this protein also requires 8mM KPVLRTA polypeptide as an additive.
[0054] The complex crystallization conditions of EV71-3C and AG7088: 0.1 M sodium acetate (pH4.6), 0.2M ammonium sulfate and 25% (w / v) PEG 4000, the molar concentration ratio of protein to compound is 1:3.
[0055] 2. Data collection and structure analysis:
[0056] Data collection uses an indoor X-ray diffractometer (in-house Rigaku MicroMax007 rotating-anode X-ray generator). Data processing uses HKL...
Embodiment 3
[0060] Example 3 The substrate binding groove of EV71-3C protease
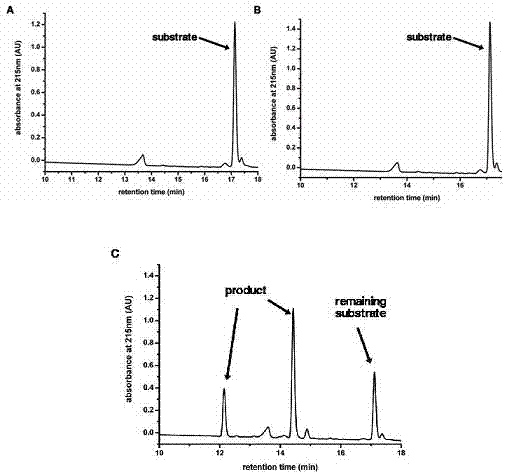
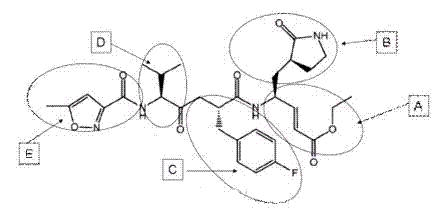
[0061] The nomenclature of each site of the 3C protease substrate binding groove: usually we use the peptide bond cleaved on the substrate polypeptide as the reference point, and the amino acids to the left are named P1, P2, P3, P4,...Pn in sequence, and the amino acids to the right are named in sequence Named P1', P2', P3', P4',...Pn'; and the part of the protease that accommodates these polypeptide amino acids is correspondingly named as the site S1, S2, S3, S4,...Sn, S1 ', S2', S3', S4',...Sn'.
[0062] Through the complex structure of compound AG7088 and EV71-3C protein, we were able to determine the substrate binding groove of EV71-3C protease ( Figure 5 ), Figure 5 The brown stick model in A is AG7088; AG7088 binds in the groove-like structure on the surface of the protease, which is the substrate binding groove of EV71-3C protease (see Figure 5 B). Through structural analysis, we found the amino a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com