Expression vector for expressing blood coagulation factor VIII and application thereof
A technology for coagulation factor and plant expression vector, which is applied to the expression vector expressing coagulation factor VIII and its application field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1. Construction method of Chlorella ellipsoid expression vector
[0054] Primers were designed according to the promoter NRpro sequence of the highly efficient nitrate reductase gene of Chlorella ellipsoides reported by Wang et al. (wang et al., 2004), and the upstream primer NRpro-f: 5'TG AAGCTT GTACCAGTGGTGCTGAGGTAGA-3' (SEQ ID NO: 4); downstream primer NRpro-r: 5'AC TCTAGA CTTGTCGTCCTACTGCCGCAACAC-3' (SEQ ID NO: 5), wherein a Hind III restriction site (underlined part) is added to the upstream primer, an Xho I restriction site (underlined part) is added to the downstream primer, and PCR Methods The NRpro promoter was obtained from the genome of Chlorella ellipsoides, and the sequencing results showed the NRpro promoter sequence (SEQ ID NO: 1). The PCR reaction conditions were: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 2 min, and extension at 72°C for 10 min after 30 cycles. The fra...
Embodiment 2
[0055] Example 2. Expression vector constructed using pGreen0029 framework.
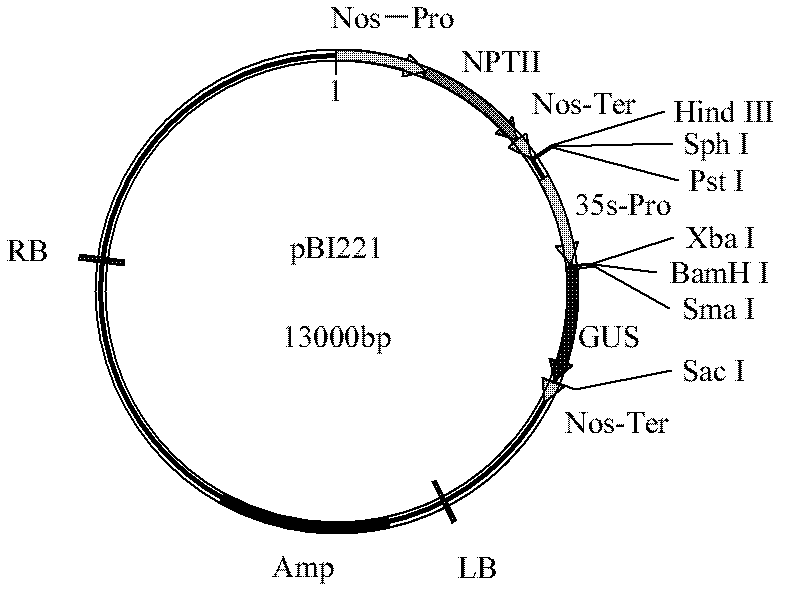
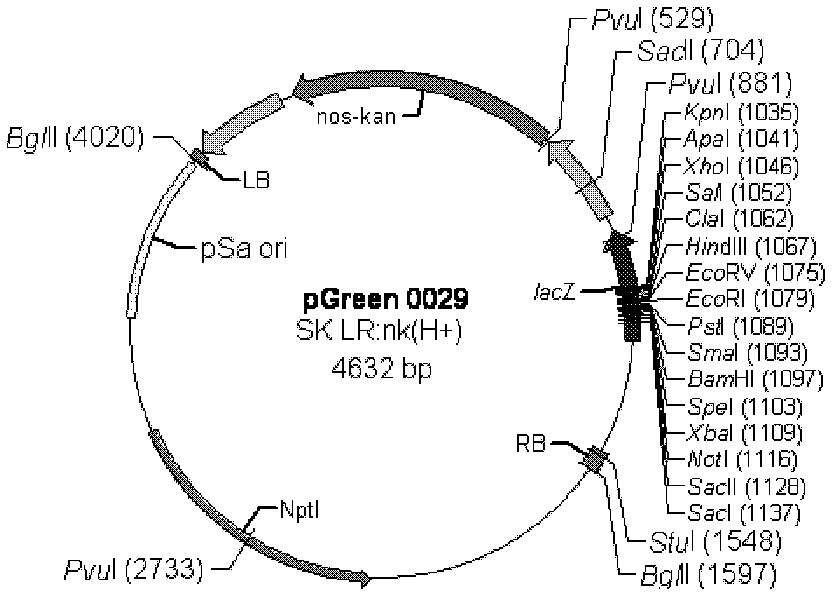
[0056] Primers were designed according to the Nos terminator sequence (SEQ ID NO: 3), upstream primer: 5'- ATAAGAATGCGGCCGC TCGAATTTCCCCGATCGTTCAAAC-3' (SEQ ID NO: 6) downstream primer: 5'- CGAGCTCG CCCGATCTAGTAACATAGATGA-3' (SEQ ID NO: 7) wherein a Not I restriction site (underlined part) is added to the upstream primer, a Sac I restriction site (underlined part) is added to the downstream primer, and PCR The method from pBI221 (from the vector of Clontech Company, the vector diagram sees figure 1 ) obtained the Nos terminator (ie figure 1 Nos-Ter in ), the PCR reaction conditions were: 94°C pre-denaturation for 3 min, 94°C denaturation for 30 s, 56°C annealing for 30 s, 72°C extension for 30 s, and after 30 cycles, 72°C extension for 10 min. The 268bp fragment of the PCR product was double-digested with Not I and Sac I endonucleases, and the plasmid pGreen0029( image 3, from BBSRC) were also...
Embodiment 3
[0057] Example 3. Synthesis of BDD-rhFVIII gene
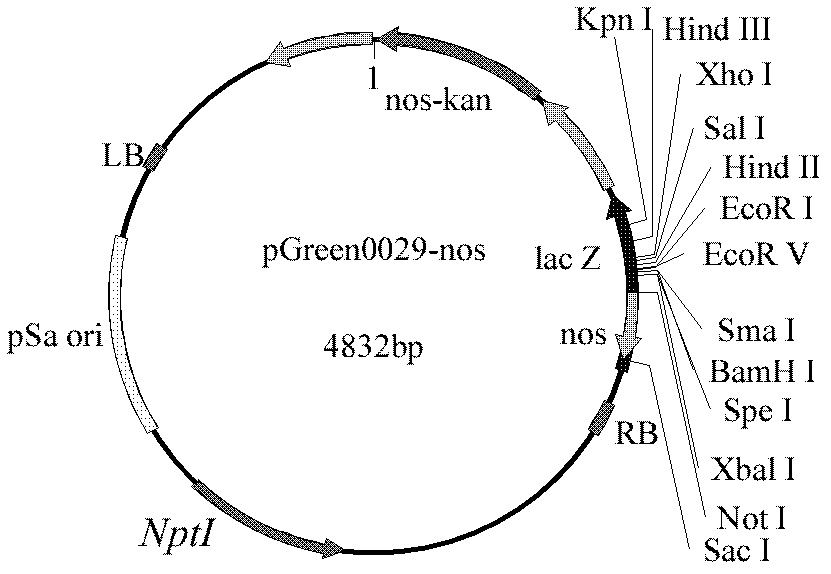
[0058] The BDD-rhFVIII gene (SEQ ID NO: 2) was synthesized by the method of gene synthesis, and double-digested by restriction sites XhoI and Not I, and a fragment of about 4.3 kb (SEQ ID NO: 2) was recovered, and then combined with Carry out double digestion vector pGreen0029-nos ( image 3 ) ligation, ligation overnight at 16°C, the ligation product was transformed into Escherichia coli DH5α, cultured upside down on LB medium containing 50 mg / L kanamycin at 37°C overnight, and the plasmids extracted from the grown resistant colonies were subjected to XhoI and Not I internalization Dicer double digestion identification, the plasmid that can obtain a fragment of about 4.3 kb (i.e. rhFVIII gene) is named pGreen0029-FVIII-nos ( Figure 5 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com