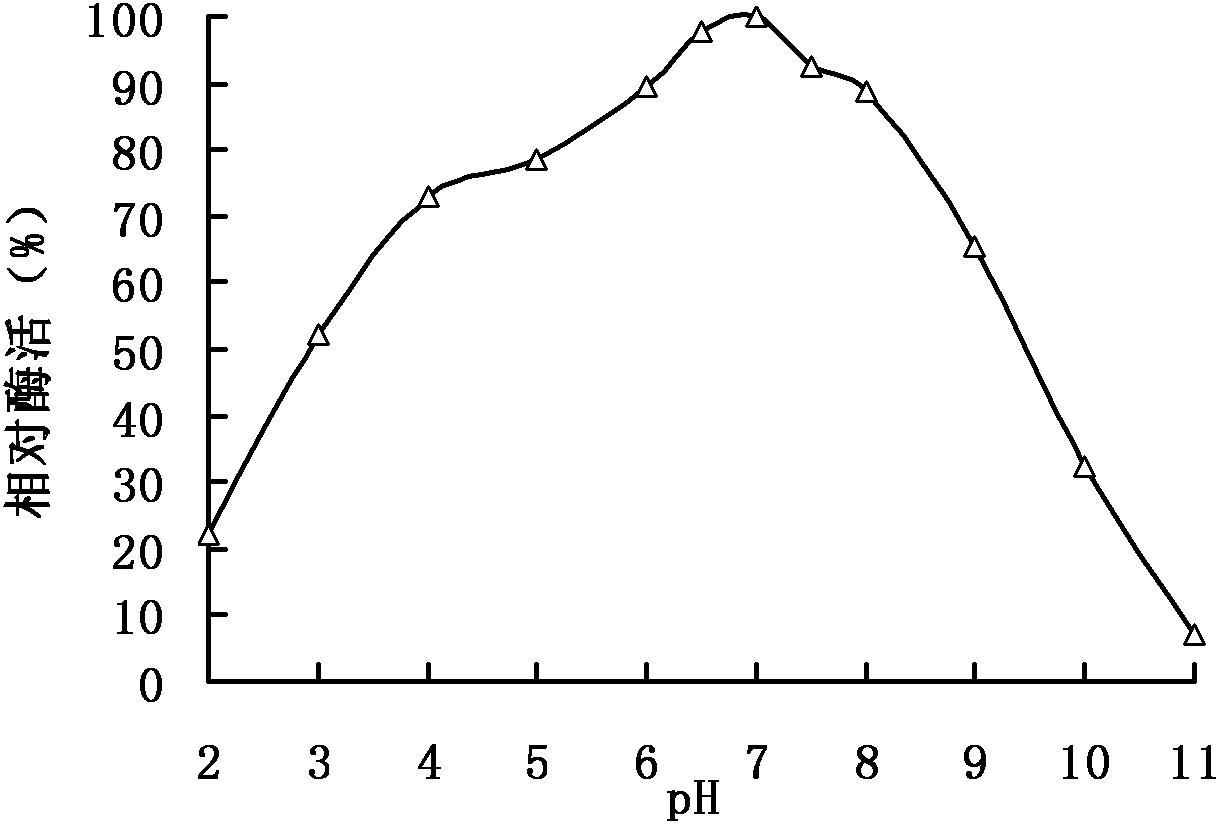
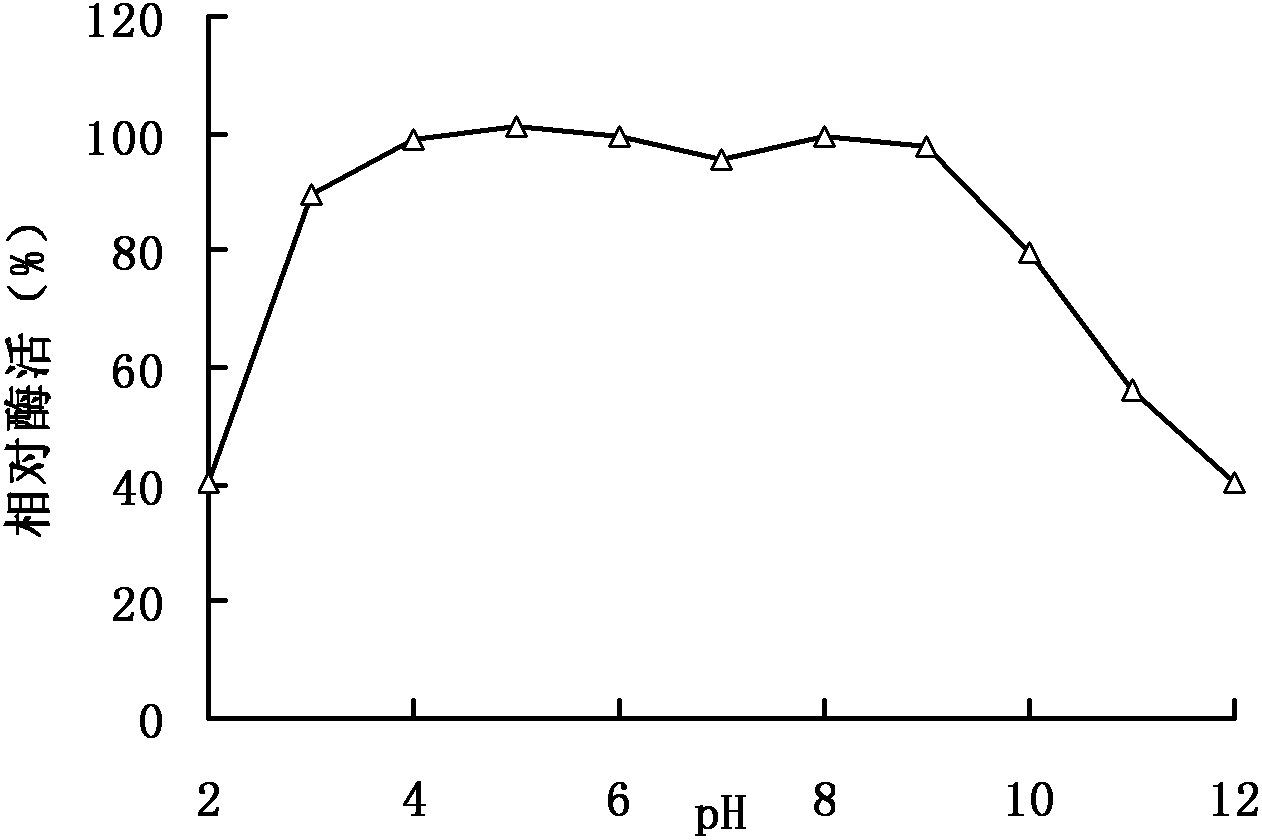
A kind of feed pectinase pla and its gene and application
A pectinase and feed technology, applied in the field of genetic engineering, can solve the problems of low expression level, inappropriate pH or temperature range, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Example 1 Extraction and purification of soil microbial metagenomic DNA
[0099] Soil samples were collected from Guangxi Juice Factory, and the metagenomic DNA of soil microorganisms was extracted by combining physical and chemical cracking methods. The physical method was mainly liquid nitrogen grinding, and the chemical cracking method was mainly based on Brady (Brady SF, Nat Protoc 2007, 2: 1297 -1305), the following is the specific operation process:
[0100] (1) Add 10g of soil to the mortar, pour liquid nitrogen into it to freeze, grind the soil vigorously with a pestle, add liquid nitrogen to grind again after thawing, and repeat three times to make the soil fully ground;
[0101] (2) Add the ground soil into a 50ml centrifuge tube, add 20ml of preheated DNA lysis buffer solution, mix it upside down and put it in a 70°C water bath for 3 hours, during which time the centrifuge tube is turned upside down every 15 minutes Second, the purpose is to fully decompose ...
Embodiment 2
[0111] Example 2 Obtaining of the Conserved Region Sequence of the Pectinase Gene
[0112] According to the conserved (A Q Y P N G G W / F and W A / G Q Q H / Y D E) sequence of the pectin lyase pf09492 gene, degenerate primers pf09492-PF and pf09492-PR were designed and synthesized:
[0113] pf09492-PF: 5'-GCTCAGTACCCNAAYGGNGGNT-3';
[0114] pf09492-PR: 5′-CTCGTCGTRYTGYTGNSCCCA-3′
[0115] Wherein: Y=C / T, R=A / G, S=C / G, N=A / T / G / C.
[0116] Using the above-mentioned degenerate primers to amplify a partial fragment of the target gene by PCR using soil organism metagenomic DNA as a template. The PCR reaction parameters are: 95°C for 5 min; 94°C for 30 sec, 51°C for 30 sec, 72°C for 1 min, after 30 cycles; 72°C for 10 min, agarose electrophoresis detection. A fragment of about 250bp was obtained, and after recovery, it was combined with pEASY-T 3 Carrier ligation was sent to Sanbo Biotechnology Co., Ltd. for sequencing.
Embodiment 3
[0117] Example 3 Obtaining of the full-length sequence of the pectinase gene
[0118] One pectinase gene fragment was randomly selected from a large number of pectinase fragments obtained above to clone the whole gene. According to the known sequence fragments of the target gene, four specific primer pairs SP1, SP2, SP3, and SP4 were designed. Each specific primer pair included a specific upstream primer and a specific downstream primer, and their sequences are shown in Table 1.
[0119] The first round of TAIL-PCR amplification was carried out on the selected pectinase gene fragments with the specific primer pair SP1. Aiming at the low content of the target sequence, a single step specific primer SP1 was added to amplify the target template sequence. to increase the copy number of the target fragment. The second-round TAIL-PCR amplification was performed on the first-round TAIL-PCR product with specific primer pair SP2 and degenerate primers. First, AD primers are added sep...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com