Method for transcriptional gene expression regulation and application
A technology of transcription level and gene expression, which is applied in the direction of introducing foreign genetic material, DNA/RNA fragments, recombinant DNA technology, etc. by using vectors, and can solve problems that do not involve gene upregulation or activation at the transcription level, and achieve powerful gene regulation functions, Effective effect, anticancer effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Design of small double-stranded RNA
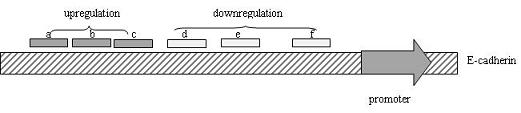
[0047] Through literature search, determine the transcription start point and core promoter region of E-cadherin gene (eg http: / / www.ncbi.nlm.nih.gov / Gene ID: 999). Log in to the website of the National Center for Biotechnology Information (http: / / www.ncbi.nlm.nih.gov / ), and download the E-cadherin gene sequence. Within the E-cadherin promoter region (1000 bp above the transcription start point), avoid high CpG regions, and select relatively complex base combinations of C, T, A and G (mainly to avoid the simple structure of the target sequence, The 19-base sequence with high repeatability) is used as the target of the small double-stranded RNA, so that the dsRNA sequence is designed according to the selected target, and the designed dsRNA is used with the blast database of the National Center for Biotechnology Information website and the whole genome For matching, select a dsRNA sequence with better specificity as the scr...
Embodiment 2
[0053] Example 2 Effects of small double-stranded RNAs on cell growth, metastasis, and invasion
[0054] Transfection of double-reporter-validated dsRNA into prostate cancer cell lines (PC-3 and DU-145) and liver cancer cell lines (97L, 97H, and LM3) to evaluate the screened cells using cell proliferation assays, scratch assays, and invasion assays Changes in biological behaviors such as growth, metastasis, and invasion of tumor cells after the action of dsRNA.
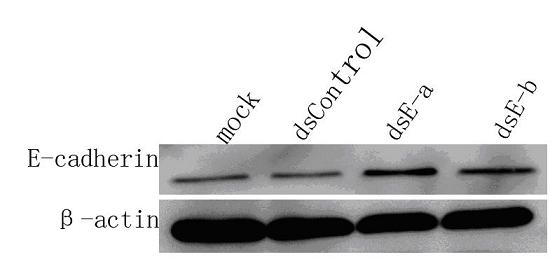
[0055] The synthetic small double-stranded RNA was transfected into prostate cancer cell lines (PC-3 and DU-145) and liver cancer cell lines (97L, 97H and LM3) using liposome-based transfection reagents, and harvested 3 days after transfection Cells, total RNA and total protein were extracted, and then the expression of the target gene was detected by fluorescent quantitative PCR and Western blotting. If the expression of the target gene is up-regulated or down-regulated after dsRNA intervention, the dsRNA will be us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com