Oligo dT primer and method for constructing cDNA library
A DNA molecule and library construction technology, applied in the field of Oligo dT primers and cDNA library construction, can solve the problems of large operation restrictions, small scope of application, and single library construction method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
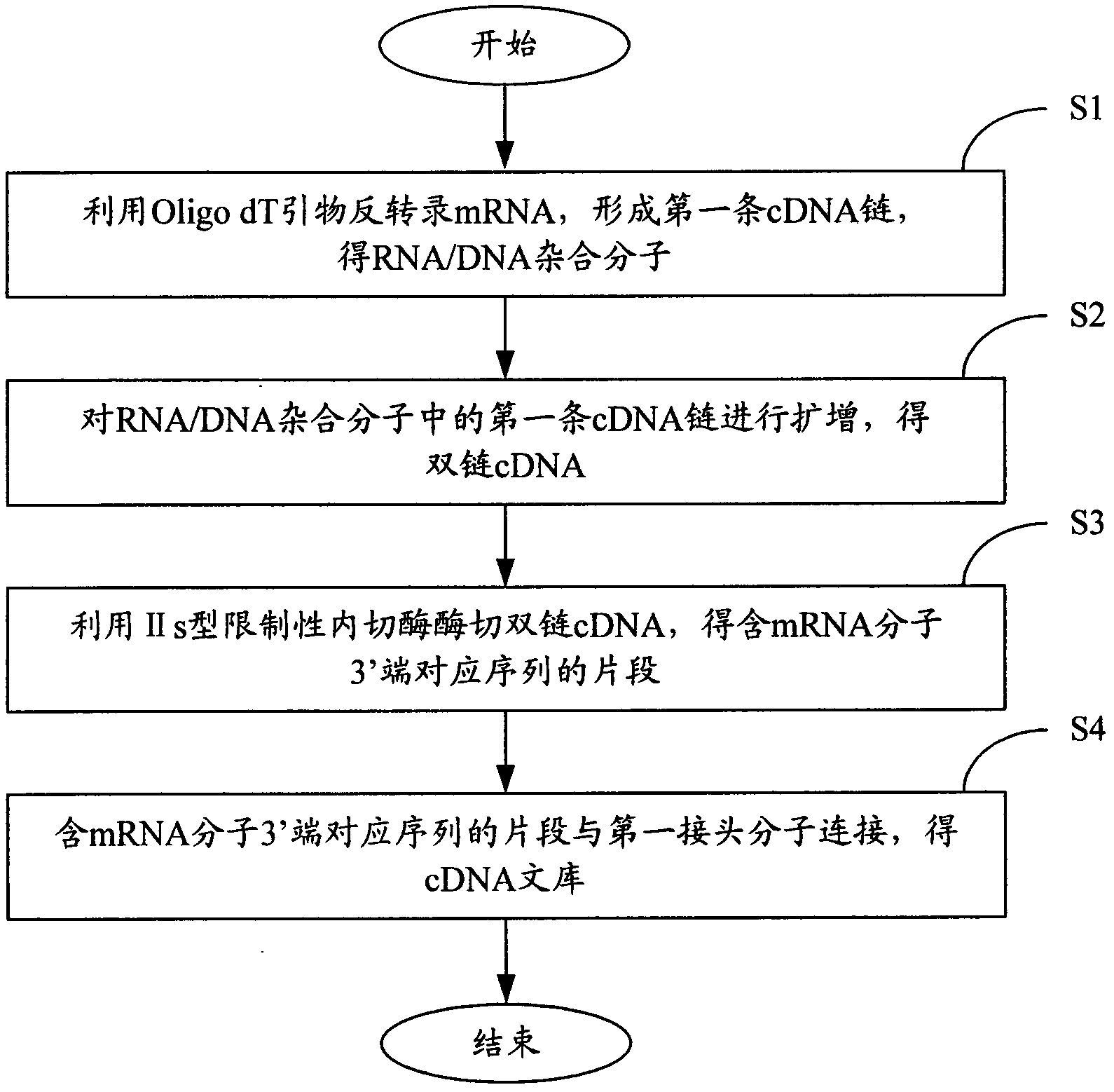
[0091] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments.
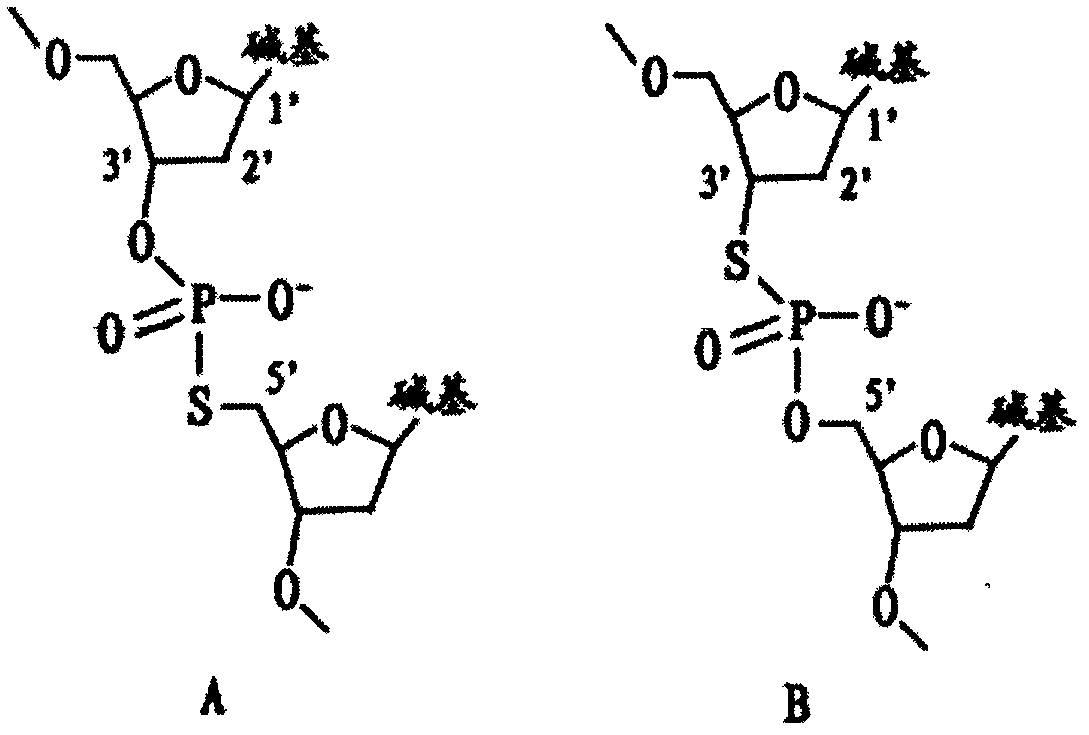
[0092]The type IIs restriction endonuclease of the present invention is a restriction endonuclease whose cutting site is outside the recognition sequence, including but not limited to: Acu I, Alw I, Bbs I, BbV I, Bcc I, BceA I , BciV I, BfuA I, Bmr I, Bpm I, BpuE I, Bsa I, BseM II, BseR I, Bsg I, BsmA I, BsmB I, BsmF I, BspCN I, BspM I, BspQ I, BtgZ I, Ear I, Eei I, EcoP15 I, Fau I, Fok I, Hga I, Hph I, HpyAV, Mbo II, Mly I, Mme I, Mnl I, NmeAIII, Ple I, Sap I, SfaN I and TspDT I, preferably Acu I, Bsg I, EcoP15 I or Mme I.
[0093] The mRNA of the present invention is derived from eukaryotic organisms, including but not limited to animals, plants, fungi and protists.
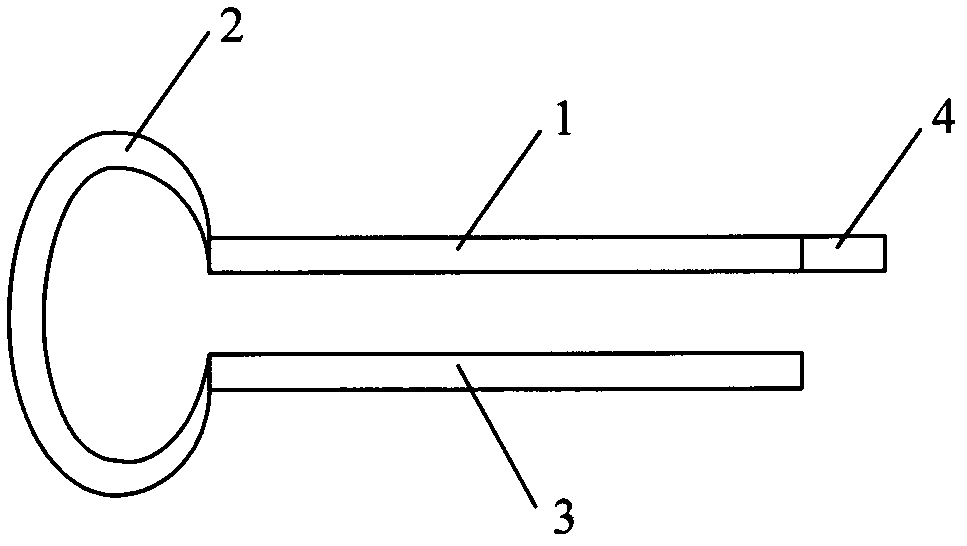
[0094] An Oligo dT primer, the Oligo dT primer is a single-stranded DNA molec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com