Method for quickly detecting microorganisms
A detection method and microbial technology, applied in the direction of color/spectral characteristic measurement, fluorescence/phosphorescence, material excitation analysis, etc., can solve problems such as insurmountable drift interference, drift of microbial growth curve, and influence on the accuracy of detection results, etc., to improve Applicability, effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] A rapid detection method for microorganisms, used to detect E. coli in water samples, comprising the following steps:
[0045] A. Setting Thresholds and Modeling Steps
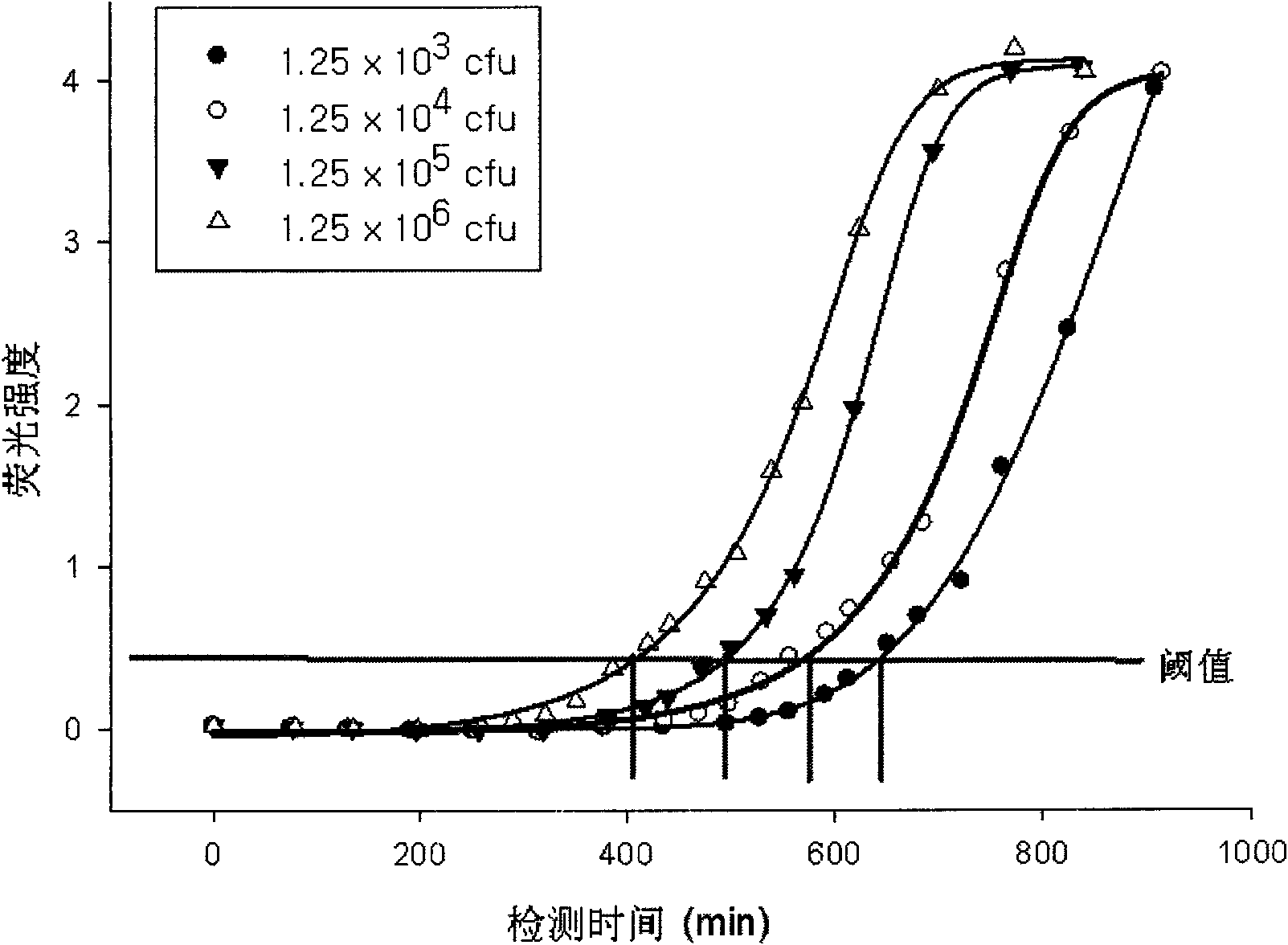
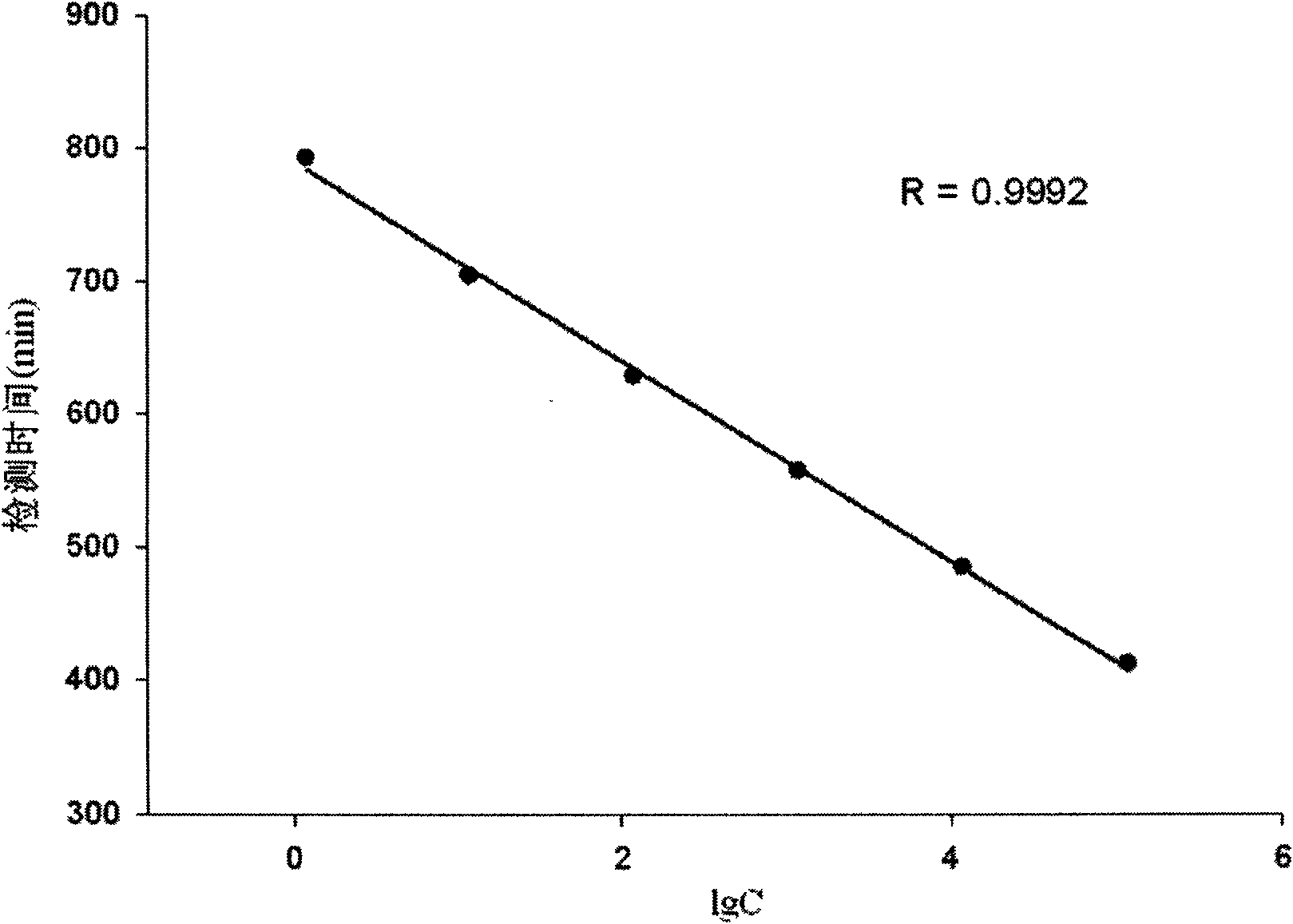
[0046] Four Escherichia coli standard samples (standard sample a, standard sample b, standard sample c and standard sample d, the concentrations C were 95.30cfu, 1.34×10 3 cfu, 1.22×10 4 cfu and 2.00×10 6 cfu) were added to the culture solution respectively, and the Escherichia coli in each standard sample grew in the culture solution respectively, and the culture solution contained the reaction substrate 4-methylumbelliferone-β-D glucuronide (MUG), and the MUG were respectively React specifically with the metabolic indicator β-D-glucuronidase produced by the growth of Escherichia coli to generate fluorescent signals (excitation wavelength is 365nm, emission wavelength is 450nm), which are detected separately to obtain the growth of each standard sample curves, such as image 3 shown.
[0047] At t...
Embodiment 2
[0054] A rapid detection method for microorganisms, used to detect total coliforms in water samples, comprising the following steps:
[0055] A. Setting Thresholds and Modeling Steps
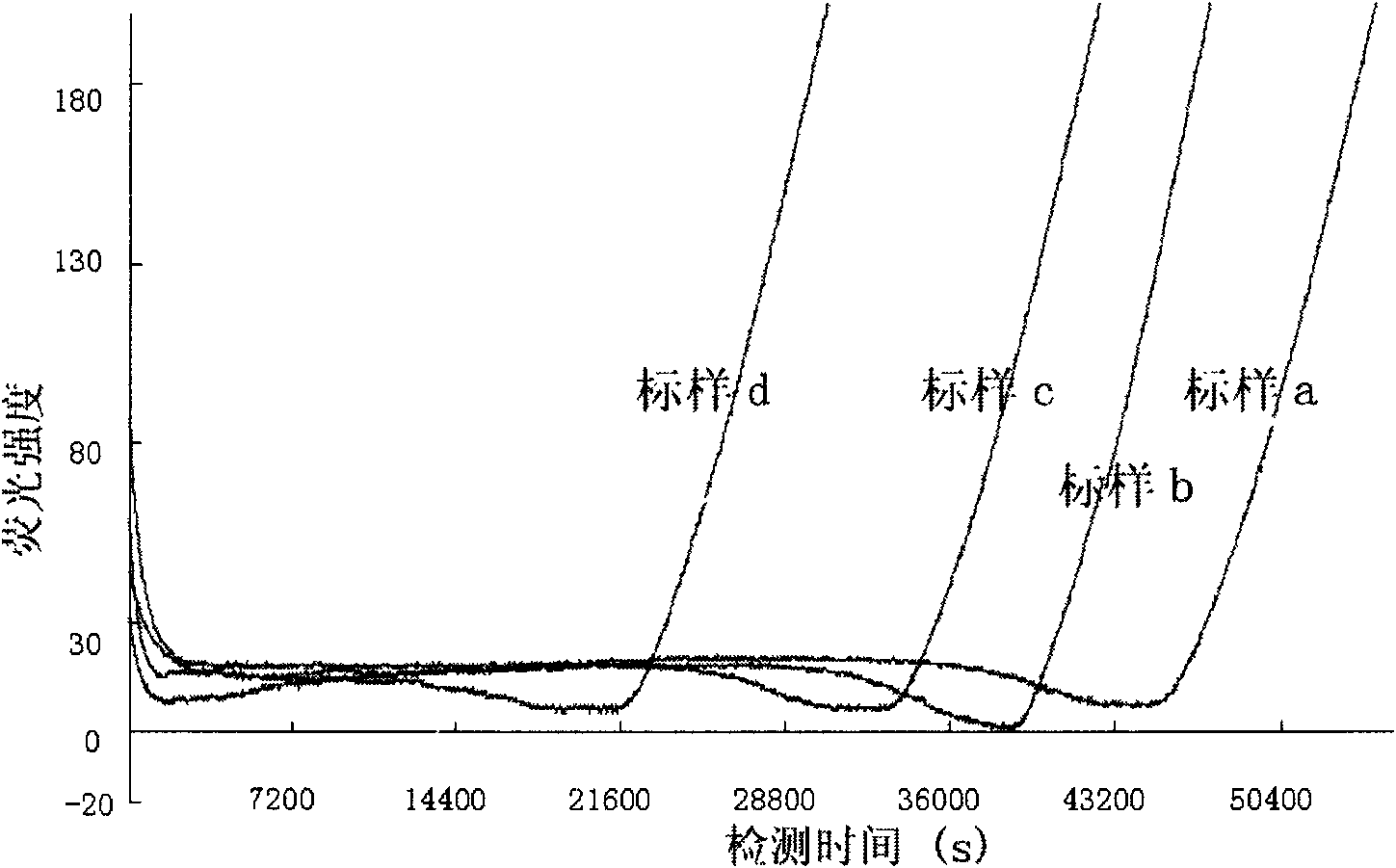
[0056] Take 4 total coliform standard samples (standard sample a, standard sample b, standard sample c and standard sample d, the concentration C of which is 95.30cfu, 1.34×10 3 cfu, 1.22×10 4 cfu and 2.00×10 6 cfu) were added to the culture solution respectively, and the total coliform bacteria in each standard sample grew in the culture solution respectively, and the culture solution contained 4-methylumbelliferone-β-D-galactocuronide (MUGal), and MUGal was respectively It reacts specifically with the metabolic indicator β-D-galactosidase produced by the growth of each total coliform group, and generates a fluorescent signal (excitation wavelength is 365nm, emission wavelength is 450nm), which are detected separately to obtain the growth of each standard sample. curves, such as Figure 4 s...
Embodiment 3
[0065] A rapid detection method for microorganisms, used to detect staphylococci in food samples, comprising the following steps:
[0066] A. Setting Thresholds and Modeling Steps
[0067] The 7 staphylococcal standard samples (standard sample 1, standard sample 2, standard sample 3, standard sample 4, standard sample 5, standard sample 6 and standard sample 7, the concentration C is 24.36cfu, 198.48cfu, 2001cfu, 14506cfu respectively , 15742cfu, 16668cfu and 1986045cfu) were added into the culture solution respectively, and the staphylococci in each standard sample grew in the culture solution respectively, and the culture solution contained reaction substrates Boc-le and u-Gly-Arp-p, respectively with each The metabolic indicator coagulase produced by the growth of Staphylococcus in the standard sample reacted specifically to produce colored p-nitroa-niline (PNA). Such a growth curve, such as Figure 5 shown.
[0068] Set the threshold value according to the slope value o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com