Nucleic acid molecular cloning method and related kit based on homologous recombination
A homologous recombination and nucleotide technology, applied in the field of DNA recombination, can solve problems such as difficulty in distinguishing single nucleotide mutations, low conversion rate, and amplification of large fragments of genomic DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0048] Preparation of extended linearized vector
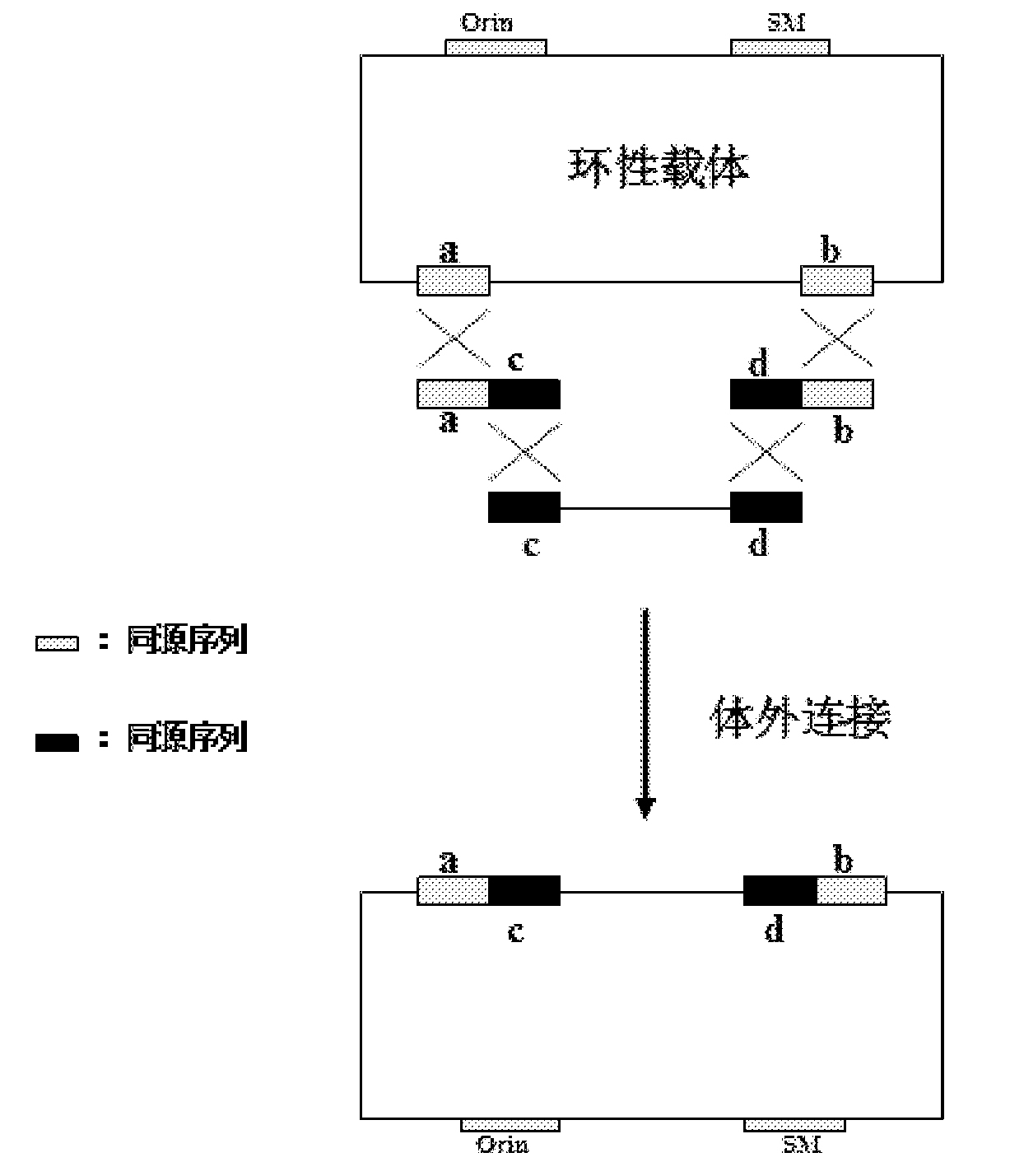
[0049] According to this application, the first sequence and the second sequence are added to both sides of the linearized vector to prepare an extended linearized vector, wherein the first sequence contains a sequence homologous to the first end of the target DNA or its flanking sequence, and the second sequence The second sequence contains a sequence homologous to the second end of the target DNA or its flanking sequence, so that the linearized vector added with the first sequence and the second sequence on both sides can be connected with the target DNA through homologous recombination. The first sequence and the second sequence may be sequences homologous to the corresponding ends of the target DNA or sequences flanking them.
[0050] The so-called "homologous" means that two nucleotide sequences have a certain sequence identity (identity) or homology (homology), so that they can be linked together by homologous recombin...
Embodiment 1
[0092] The human DHRS4 gene cluster has three gene copies, namely DHRS4 (15.569bp), DHRS4L2 (about 35kb) and DHRS4L1 (also known as DHRS4X), among which the former two are highly homologous (90%-98%) and belong to fragments Replication (segmental duplication). The homology between DHRS4L1 and DHRS4 and DHRS4L2 was 77.8% and 77.7%, respectively. The high homology between the three genes limits the application of conventional molecular biology methods and brings difficulties to the sequencing of the DHRS4 gene (full length 15.569bp), that is, through the next-generation gene sequencing technology and gene chip capture technology (from Agilent and Nalgene) are difficult to carry out accurate sequencing and SNP research of this gene.
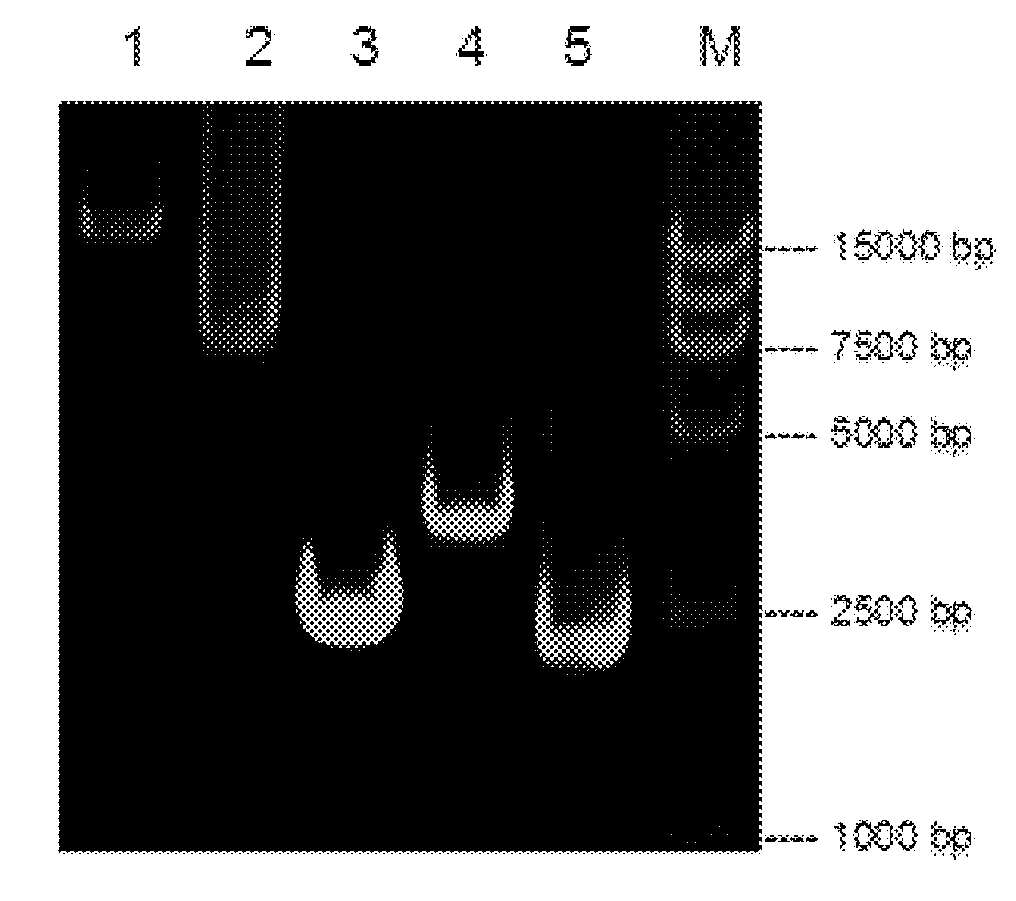
[0093] In this example, the homologous recombination of the DHRS4 gene into the p15A vector (Prutin Biotechnology (Beijing) Co., Ltd.) was mainly performed using an enzyme mixture containing RecE and RecT. In short, by using the 15-50bp sequences ...
Embodiment 2
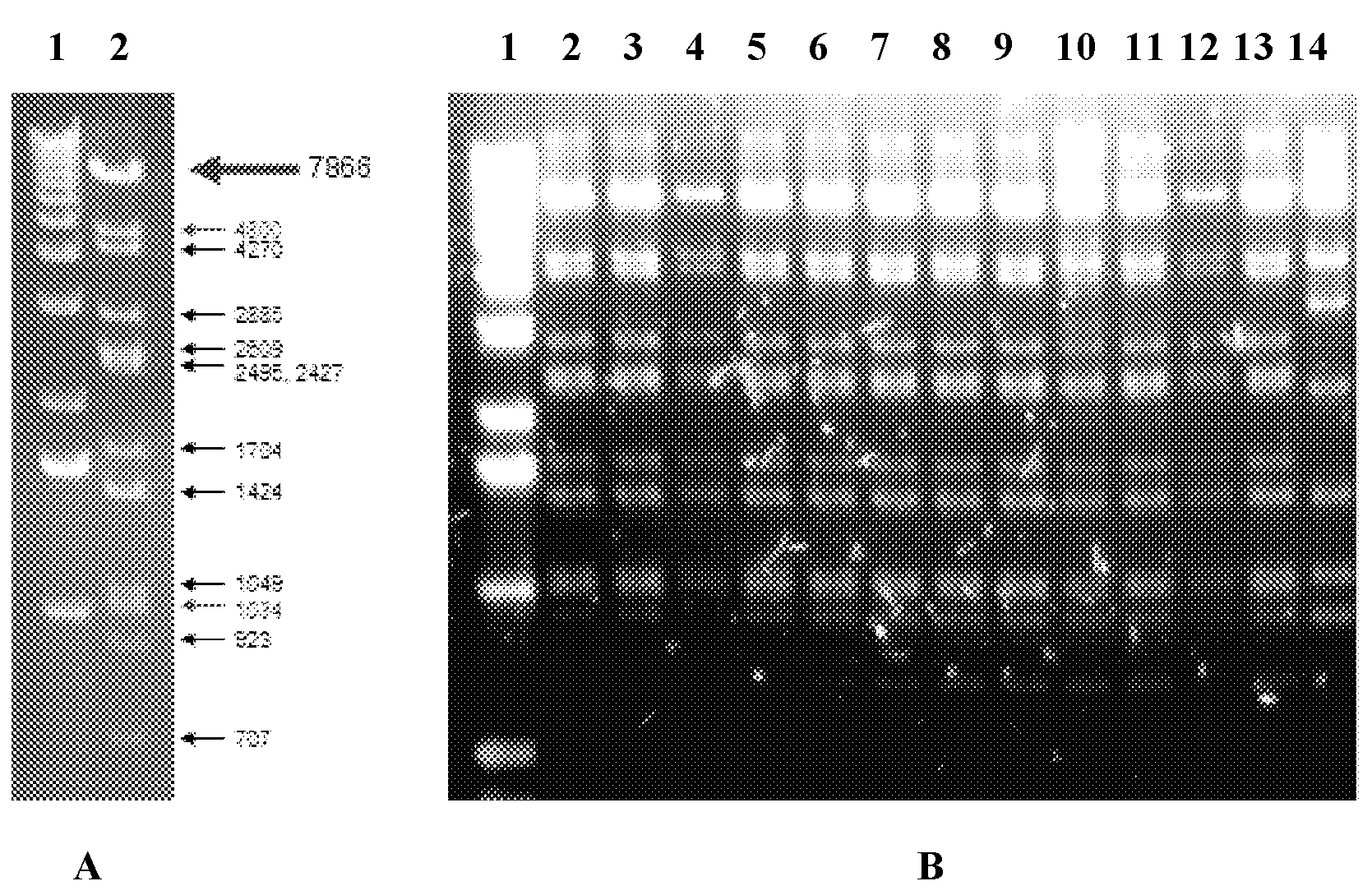
[0122] According to the method described in Example 1, suitable PCR primers were designed, and the mouse TFIIA gene (transcription factor IIA, Transcription factor II A) with a length of about 30 kb was cloned from the cosmid LAWRIST7-mTFIIA (Gene Bridges GmbH) onto the p15A vector . The PCR products of the plasmids obtained from positive clones were verified by PstI restriction map. Figure 4 The enzyme digestion patterns of the PCR products obtained from some positive clones are shown. It can be seen from the figure that lanes 2-13 in Figure B are normal connections, while lane 14 is incorrect connections. According to statistics, the successful connection rate of cloning according to the method of the present application (the number of correctly connected clones / the number of detected positive clones) is about 65-70%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com