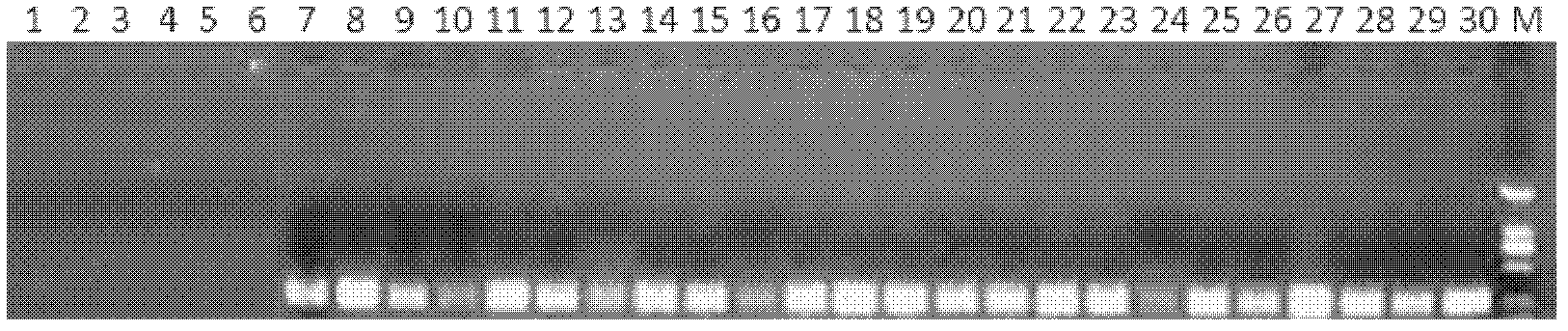
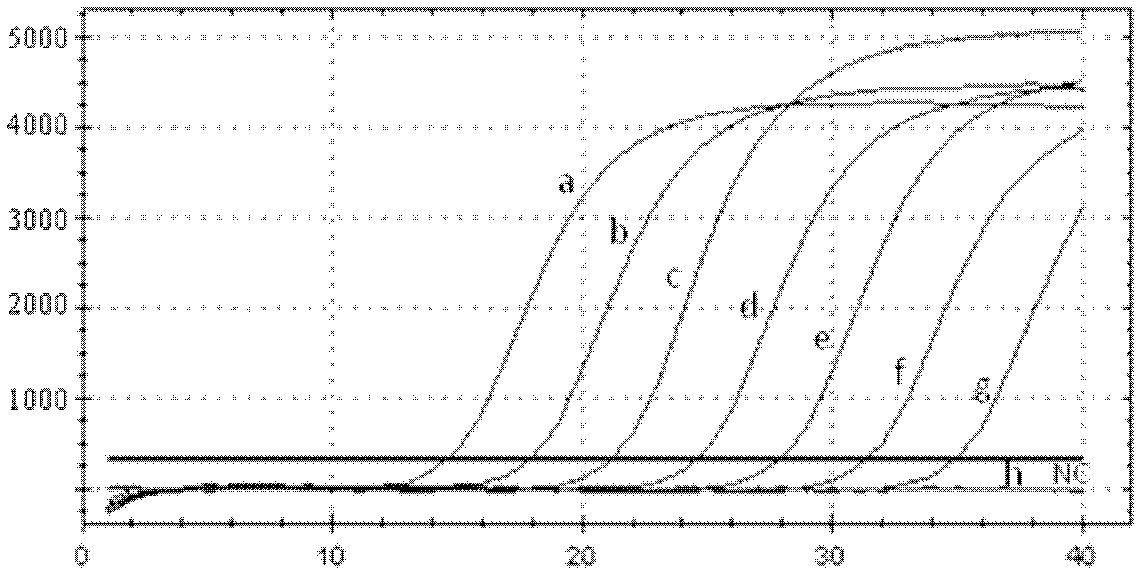
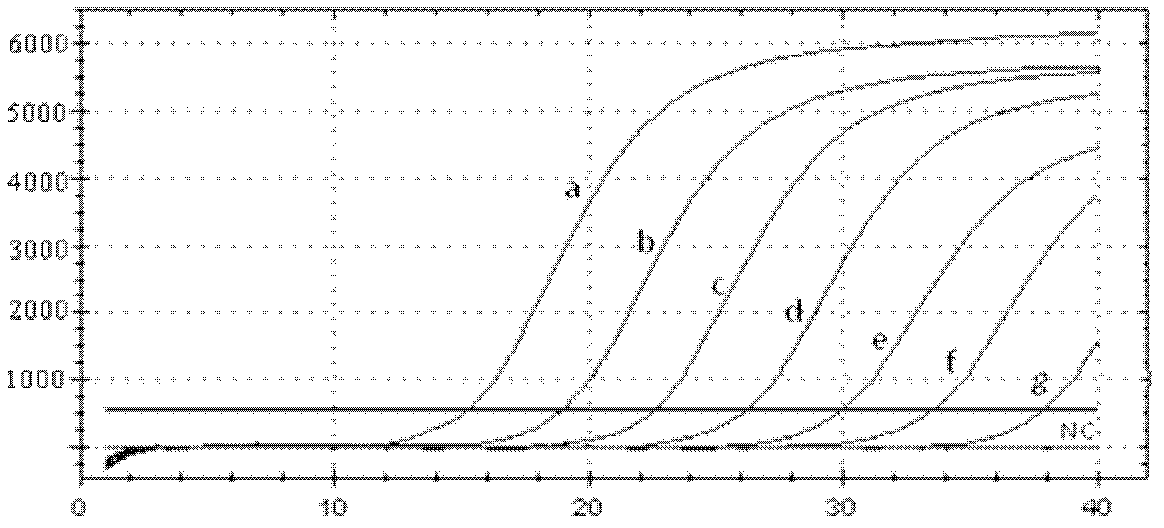
Primer for detecting reverse transcription fluorescence PCR (polymerase chain reaction) of salmonella paratyphi A
A technology for paratyphoid and Salmonella, which is applied in the field of primers for reverse transcription fluorescent PCR detection, can solve the problems of unfavorable early diagnosis, low positive rate, time-consuming H-a antigen, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0014] The design and detection method of the reverse transcription fluorescent PCR detection primer of embodiment Salmonella paratyphi A
[0015] 1 Materials and methods
[0016] 1.1 Experimental strains
[0017] 44 strains of Salmonella paratyphi used in the present embodiment, Salmonella typhi, Salmonella typhi B and C and other 31 non-typhoidal Salmonella serotypes total 57 strains (table 1), all serotypes Serotyping was performed using Salmonella antiserum (purchased from Statens Serum Institut, SSI, Denmark). In addition, other common intestinal pathogens and Streptococcus pneumoniae, Borrelia burgdorferi, Leptospira, Legionella pneumophila, Neisseria meningitidis, Rickettsia, Brucella, etc. Bacteria and other bacterial strains (Table 1) were used for specificity and sensitivity evaluation, and the above-mentioned bacterial strains were all from the Institute of Infectious Disease Prevention and Control, Chinese Center for Disease Control and Prevention.
[0018] 1.2 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com