Ivermectin and application of derivative thereof
A technology of ivermectin and its derivatives, which is applied in the field of application of ivermectin and its derivatives, and can solve problems such as ivermectin that have not been reported yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0131] Example 1. It is proved that ivermectin is a new FXR ligand with high affinity and high specificity.
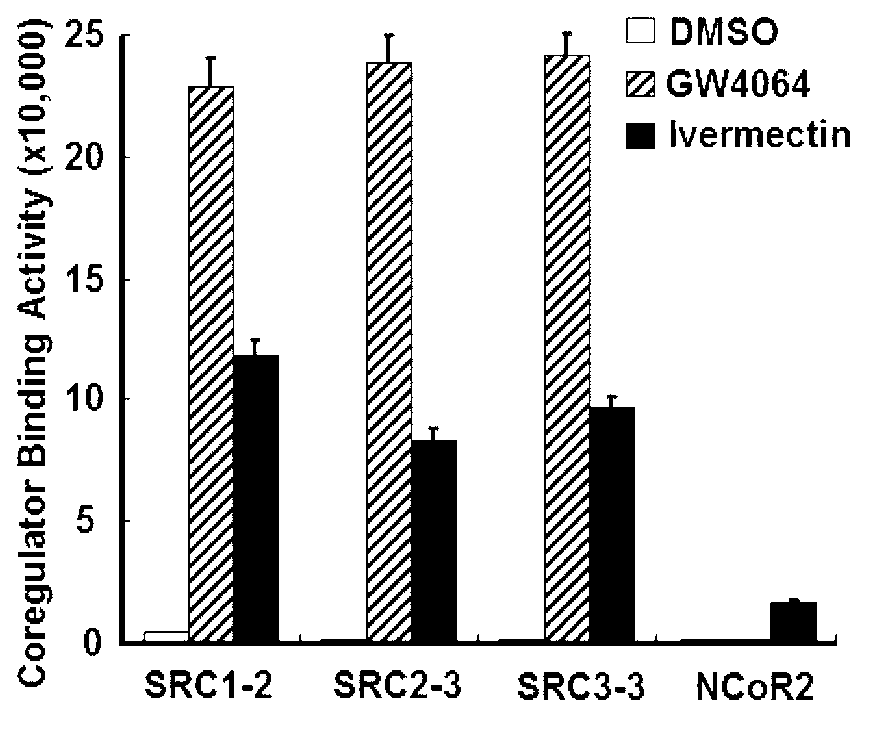
[0132] Screen the specific ligands of FXR through AlphaScreen, and detect the effect of FXR and co-activator or co-inhibitor. The test found that ivermectin can induce FXR to recruit co-activators, such as SRC1, SRC2 and SRC3, which are slightly lower than the ligand GW4064. Compared with GW4064, FXR also has a certain ability to recruit co-inhibitors ( figure 1 ). Therefore, ivermectin, as a partial activator of FXR, exerts its regulatory function in a unique way.
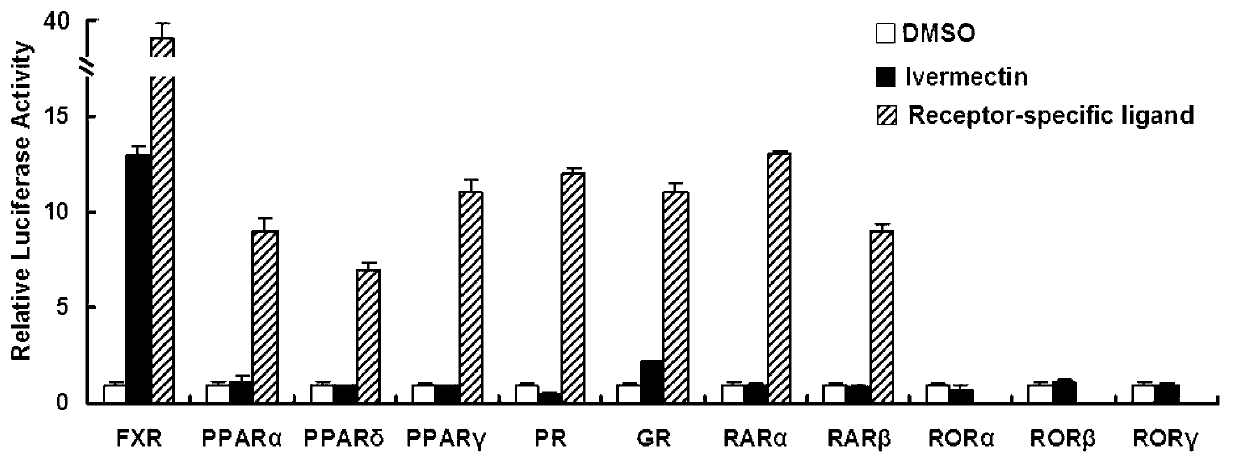
[0133] In order to prove that ivermectin can specifically bind to FXR, we co-transfected COS-7 cells with the endogenous reporter gene EcRE of FXR and a plasmid expressing full-length FXR. It was found that among the different nuclear receptors tested, ivermectin only specifically activated the transcriptional activity of FXR ( figure 2 ). This reflects the high specificity of ivermectin as FXR ligand.
[013...
Embodiment 2
[0135] Example 2. Crystal structure analysis of FXR / ivermectin complex
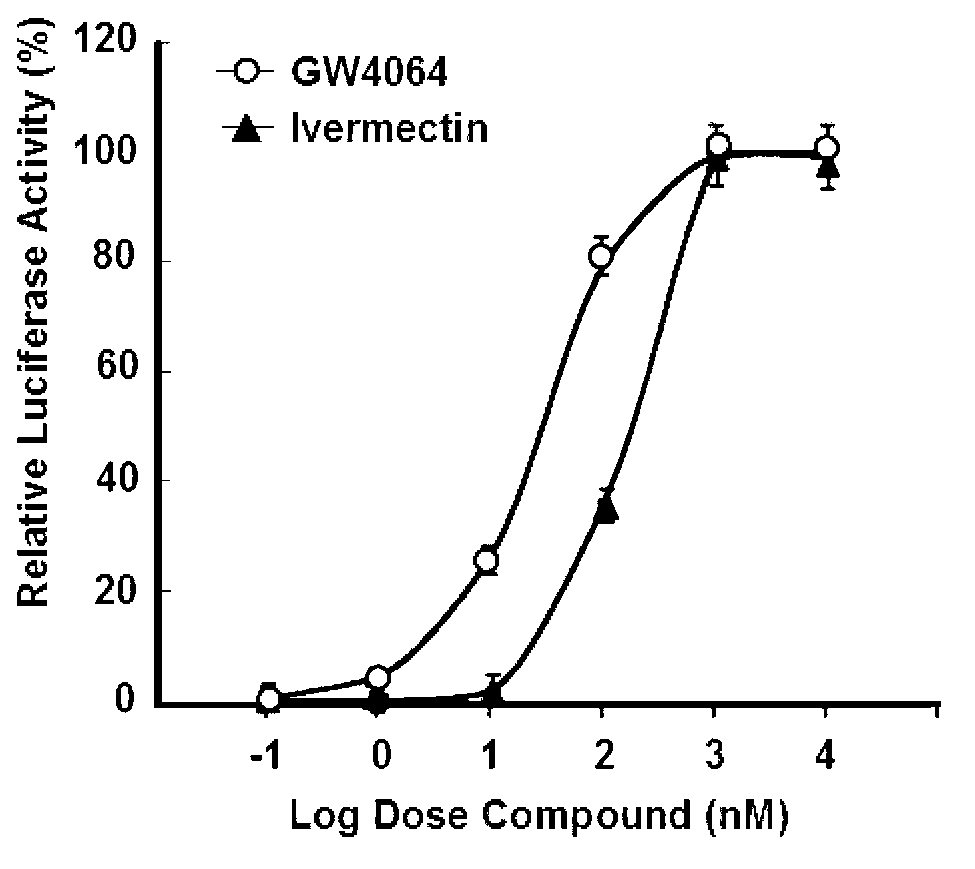
[0136] In order to reveal the molecular mechanism of ivermectin and FXR mutual recognition and binding at the molecular level, we analyzed the resolution to The crystal structure of the complex between FXR / ivermectin and the co-inhibitor NcoR nuclear receptor binding motif (Appendix 1, Figure 4 ). The structure shows that the combination of ivermectin and FXR ligand binding domain conforms to the classic "sandwich" conformation. From the electron cloud picture, ivermectin clearly exists with the ligand binding pocket of FXR ( Figure 5 ). In the structure of the complex, the electron density map cannot clearly see the helical structure of AF-2 at the carboxyl end of FXR, which is caused by the instability of the AF-2 helix due to the binding of ivermectin ( Figure 4 ). This explains from the molecular mechanism that ivermectin is weaker than the synthetic FXR ligand GW4064 in terms of inducing FXR transc...
Embodiment 3
[0137] Example 3. The unique binding site of Ivermectin in the FXR ligand binding pocket
[0138] In order to verify the amino acid sites in the FXR ligand binding pocket that directly bind to ivermectin, we made point mutations to the key sites of the FXR and ivermectin complex based on the structure of the FXR and ivermectin complex, and detected the regulation of the FXR ligand after these mutations. To determine the importance of these binding sites for ivermectin binding.
[0139] A291 plays an important role in regulating the size of the FXR ligand binding domain. When the alanine (A) at position 291 is mutated to tryptophan (W), the side chain group of tryptophan, which is much larger than alanine, occupies the space of the ligand binding pocket, which will cause FXR The ligand binding pocket will become smaller, thereby affecting the binding of FXR ligand. Consistent with our expectations, the results of the reporter gene analysis experiment showed that the mutation of A2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com