Protein interaction detection method with low false positive rate
A technology of false positive rate and detection method, which is applied in the field of molecular biology, can solve the problems affecting the application of BiFC technology and the difficulty of controlling the ratio of different gene expression, and achieve the effect of simple operation, reducing false positive and expanding the scope of application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
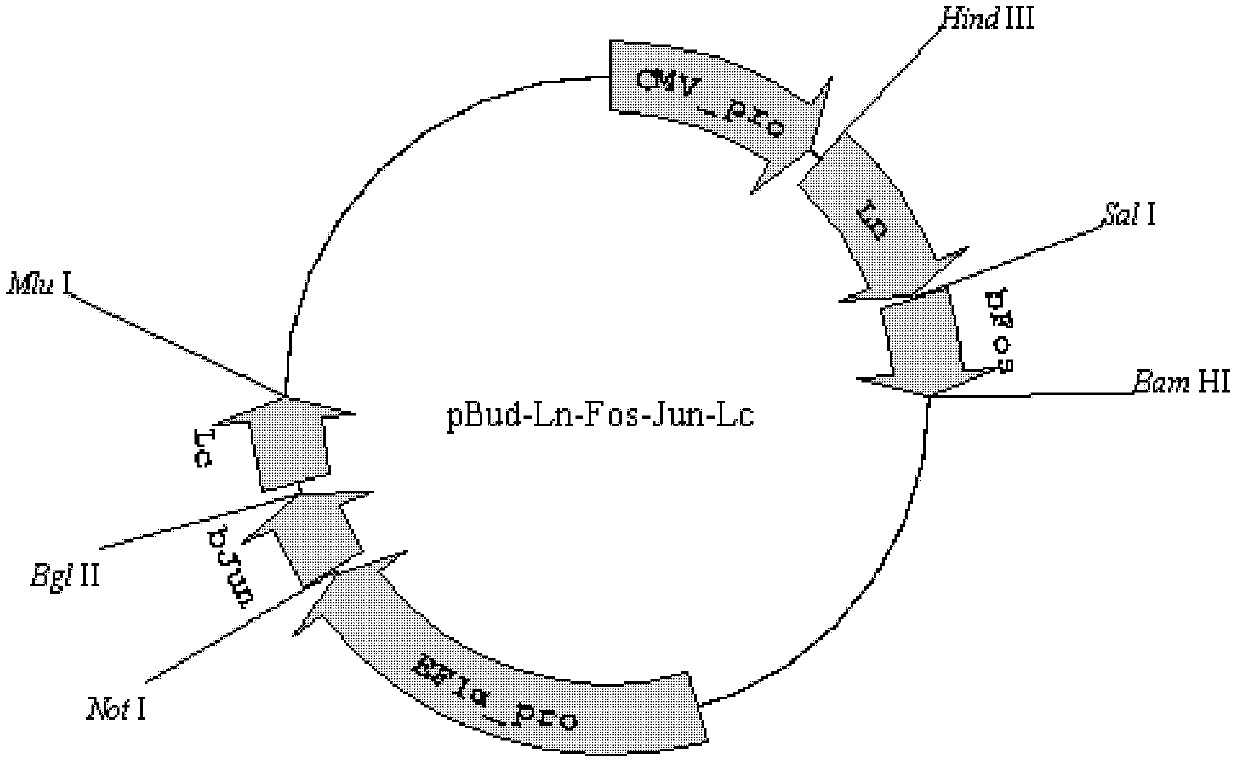
[0044] This implementation example uses the mLumin-BiFC system based on dual expression vectors to establish a protein interaction detection method with a low false positive rate. Based on this purpose, bFos was connected to the N-terminal of the mLumin protein sequence (hereinafter referred to as LN), and bJun was connected to the C-terminal of the mLumin protein sequence (hereinafter referred to as LC).
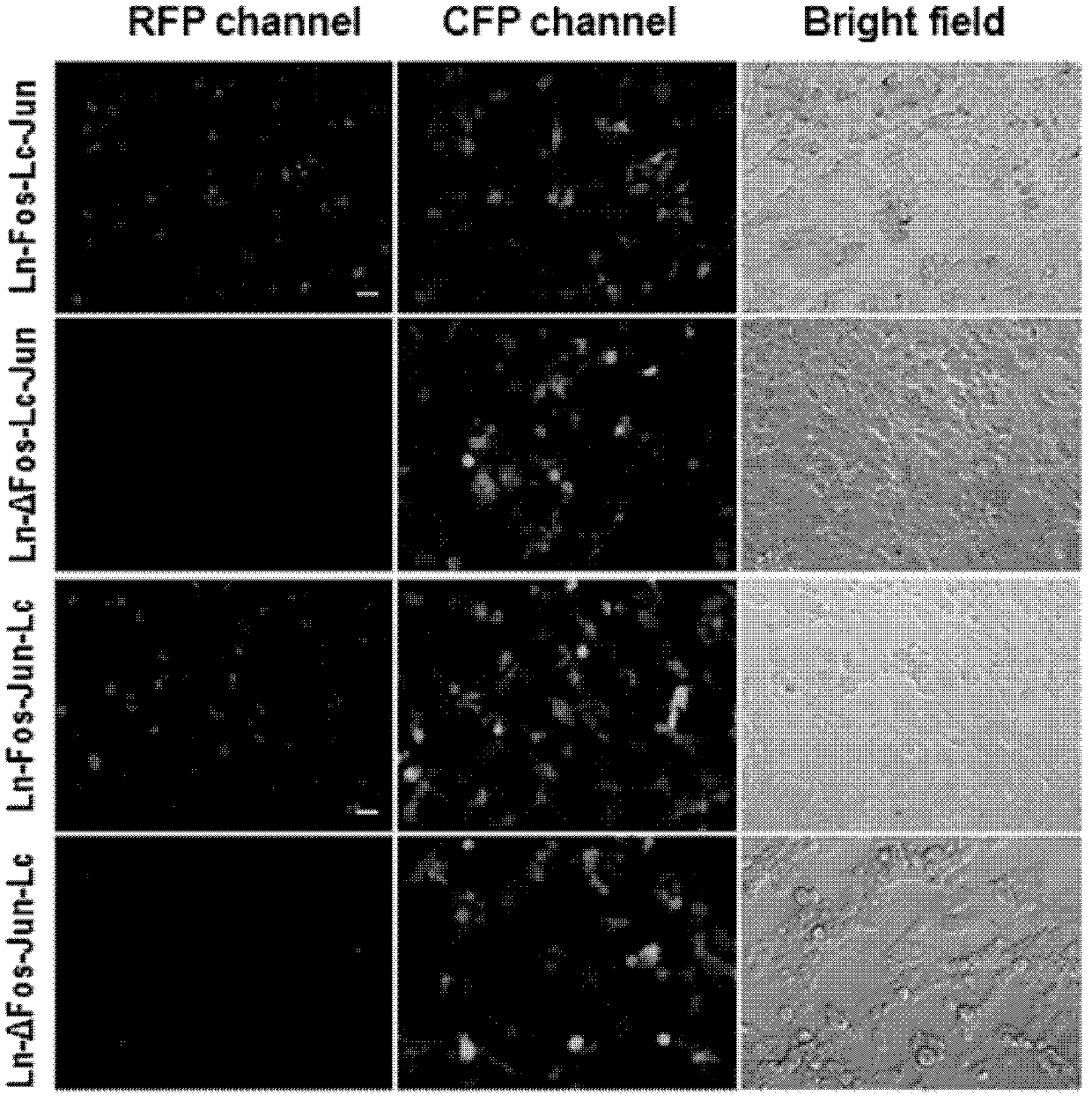
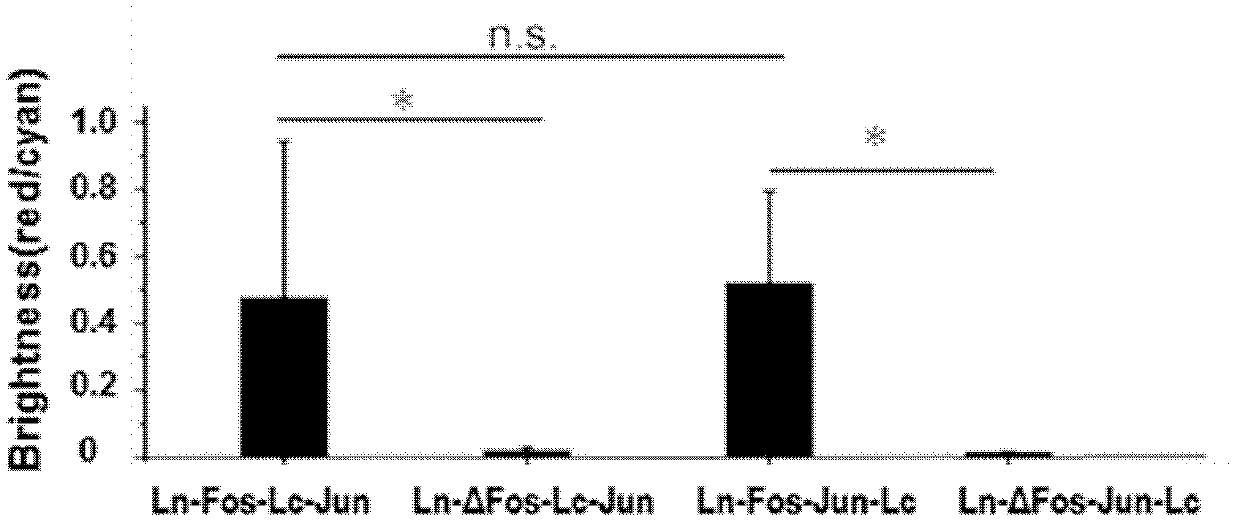
[0045] refer to figure 1 , figure 1 It is the structural diagram of the BiFC molecular probe based on the mLumin155 site, which consists of the positive group mLumin-Ln155-bFos / mLumin-Lc155-bJun and the negative group mLumin-Ln55-ΔbFos / mLumin-Lc155-bJun of the probe. Among them, ΔbFos is the bFos after deletion mutation, which cannot interact with bJun under physiological conditions, and thus does not generate BiFC fluorescence signal.
[0046] The specific steps are as follows: firstly, pBud-Ln-Lc is constructed by applying genetic engineering technology, and the plasmid...
Embodiment 2
[0051] Raf1 is the main effector protein of KRas, an important component of cell signaling pathway, and plays an important role in growth, development and carcinogenesis. The Ras-binding domain RBD of Kras-Raf1 protein has been used as a specific probe of Ras activity in many studies. GTP-bound KRas has a stronger affinity for RBD than GDP-bound KRas, and the affinity between GDP-bound Ras and Raf1 is too weak to participate in signal transduction. Therefore, heterodimerization of RBD with KRas may serve as a good protein interaction model for the development of PPI detection methods in living cells.
[0052] Figure 8 to Figure 10 It is the fluorescence map of COS-7 cells transfected with pBud-Ln-RBD-Lc-KRas, pBud-Ln-RBD-Lc-KRas 12v and pBud-Ln-RBD-Lc-KrasC185S. RBD and KRas or KRas mutants were inserted into the pBud-Ln-Lc vector to construct three groups of pBud-Ln-RBD-Lc-KRas, pBud-Ln-RBD-Lc-KRas12v and pBud-Ln-RBD-Lc-KrasC185S probe. After the COS-7 cells were transfe...
Embodiment 3
[0054] Grb2 is an important adapter protein in the cell signaling pathway. Through Grb2, KRas can interact with different signaling pathway downstream proteins. The activation of KRas is mainly accomplished through its interaction with the Grb2-SOS1 protein complex.
[0055] The fusion fragments Grb2 and KRas or their mutants were cloned into pBud-Ln-Lc to construct pBud-Ln-Grb2-Lc-KRas, pBud-Ln-Grb2-Lc-KRas 12v and pBud-Ln-Grb2-Lc-KRas185S . After the plasmids were transfected into COS-7 cells, they were cultured at 37°C for 24 hours before imaging. refer to Figure 11 to Figure 13 , the red fluorescence of cells expressing pBud-Ln-Grb2-Lc-KRas12v on the cell membrane was significantly stronger than that of pBud-Ln-Grb2-Lc-KRas, but no obvious fluorescence was seen in pBud-Ln-Grb2-Lc-KRas185S The fluorescence indicates that the BiFC probe can be applied to the detection of complexes formed by more than two proteins, and also verifies that the activation of KRas enhances th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com