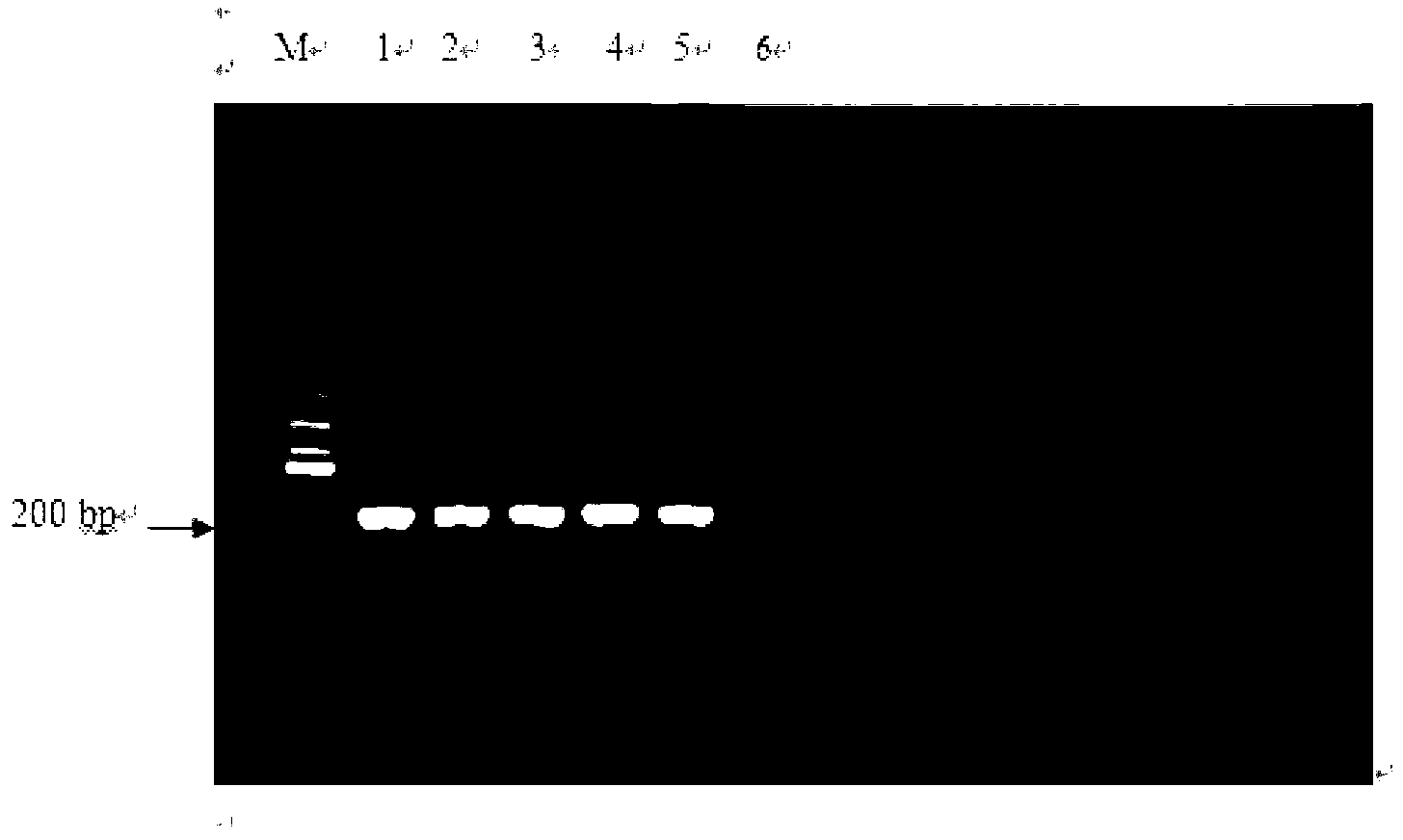Method for identifying wild strain and vaccine strain of hog cholera virus
A technology of swine fever virus and wild strains, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection, which can solve the limitation of popularization and application of fluorescent quantitative PCR detection methods, and cannot achieve high-throughput detection, not suitable for rapid identification and other problems, to achieve the effect of shortening the detection time, fast detection speed and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] In order to verify the feasibility and reliability of the method of the present invention, a standard plasmid was constructed, each plasmid sequence was saved (correctly sequenced), and then the saved plasmid sequence was amplified for HRM analysis as a standard control.
[0064] (1) Construction of standard plasmid samples:
[0065] ① Take four wild strains identified as CSFV by sequencing (two strains are from a pig farm in Shanxi named D7, T144, and the other two strains are from a pig farm in Zhanjiang, Guangdong named Z1, Z2) and a vaccine strain ( The RNA of the cell seedlings produced by Guangdong Yongshun Biopharmaceutical Co., Ltd. (HCLV strain) was reverse transcribed into cDNA;
[0066] ②Amplify and purify the cDNA obtained by reverse transcription by PCR;
[0067] ③ Ligate the purified cDNA amplification products into the pMD-18T vector with a kit from TAKARA Company,
[0068] The carrier ligation reaction system (10 μl) is as follows:
[0069] ...
Embodiment 2
[0085] HRM detection of PCR products in clinical samples
[0086] (1) Extraction of CSFV RNA from disease samples:
[0087] Use the kit MiniBEST Viral RNA / DNA Extraction Kit Ver.4.0 to extract the CSFV RNA in the samples of the disease materials, the samples are blood, lymph nodes, liver, spleen, kidney and other tissues (the wild virus samples come from a pig farm in Shanxi and Guangdong , the vaccine strain was produced by Yongshun Biopharmaceuticals in Guangdong Province).
[0088] (2) One-step RT-PCR amplification of classical swine fever virus:
[0089] The extracted CSFV RNA was amplified by one-step RT-PCR using the PrimeScript one step RT-PCR Kit Ver.2 kit (including PrimeScript 1 step Enzyme Mix, 2×1 Step Buffer, etc.).
[0090] The reaction system is as follows: use the RNA extracted in (1) as a template, add 1-5 μl of template according to the actual concentration, add 0.5 μl of PrimeScript 1 step Enzyme Mix, add 0.5 μl of upstream and downstream primers P1 and P2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com