Method for separating two tightly linked rice sterile genes
A gene and linkage technology, applied in the field of separating two closely linked rice sterility genes, can solve problems such as being susceptible to external environmental influences, research difficulties, unstable fertility, etc., and achieve the effect of eliminating the influence of genetic background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
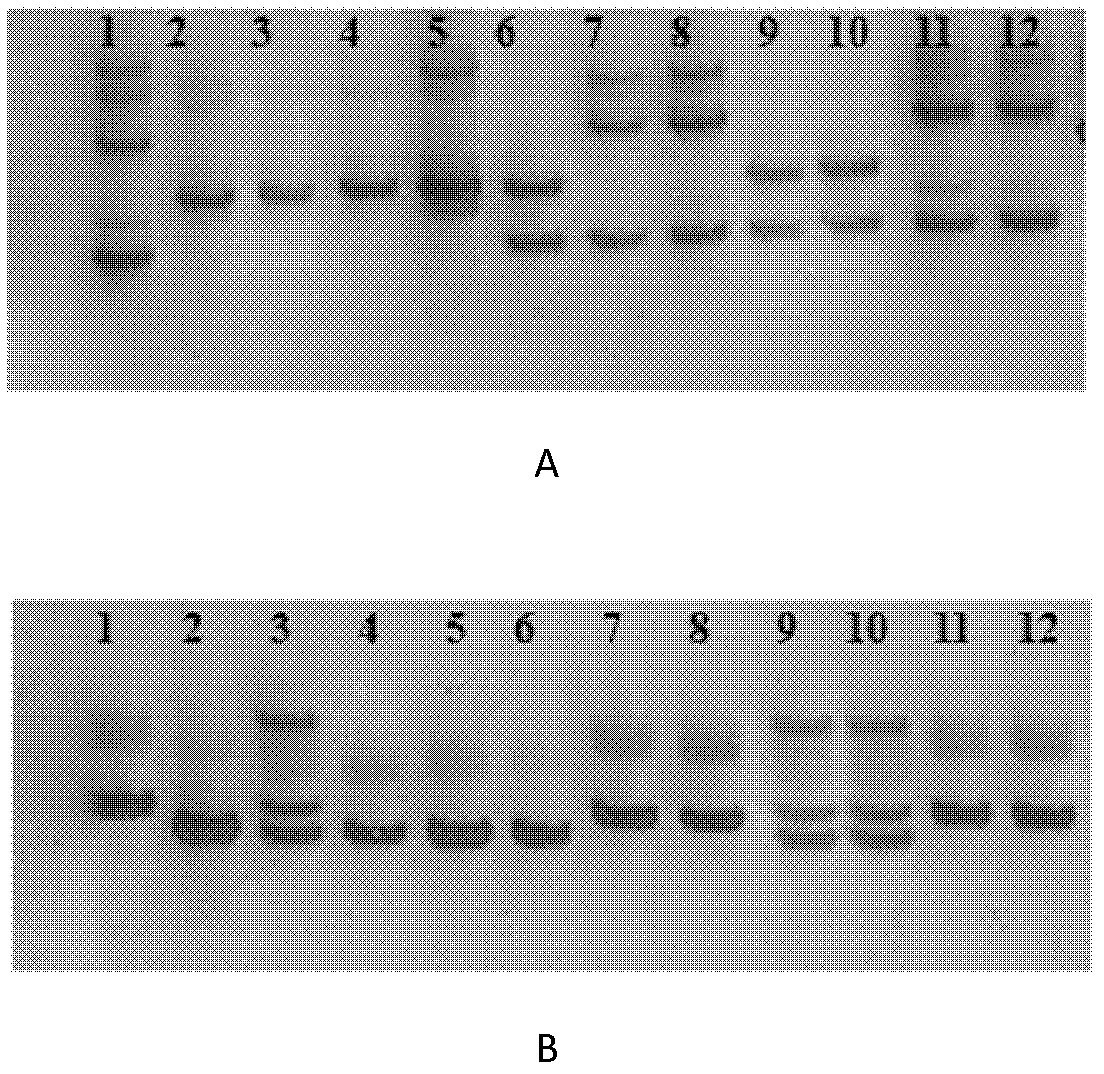
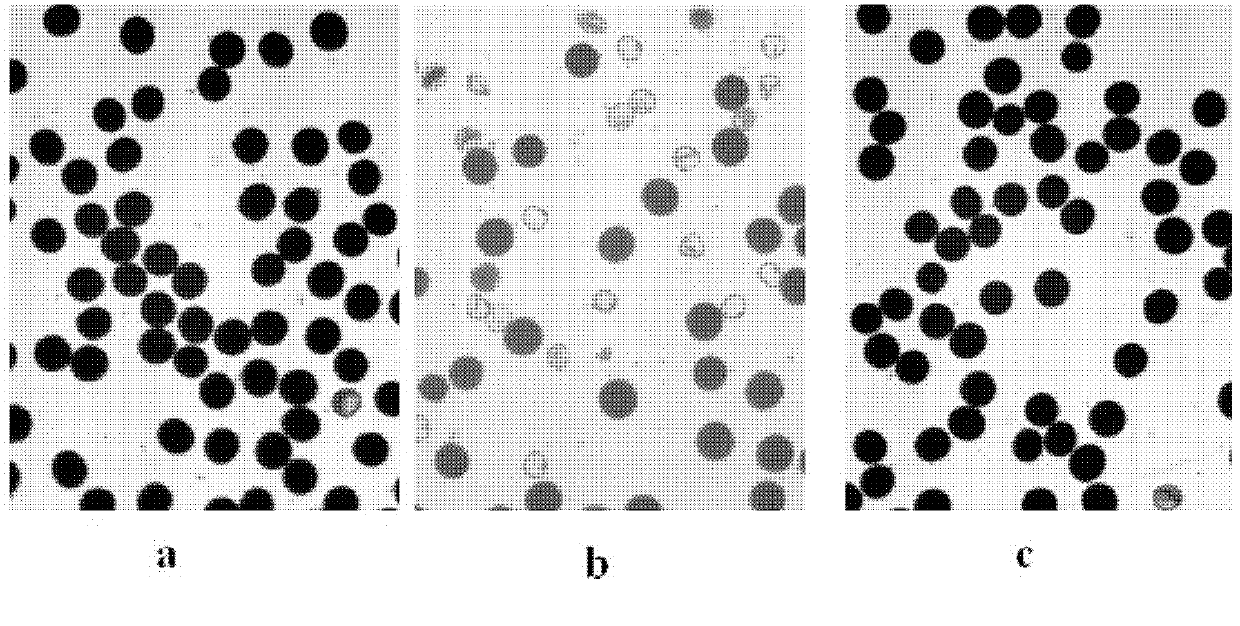
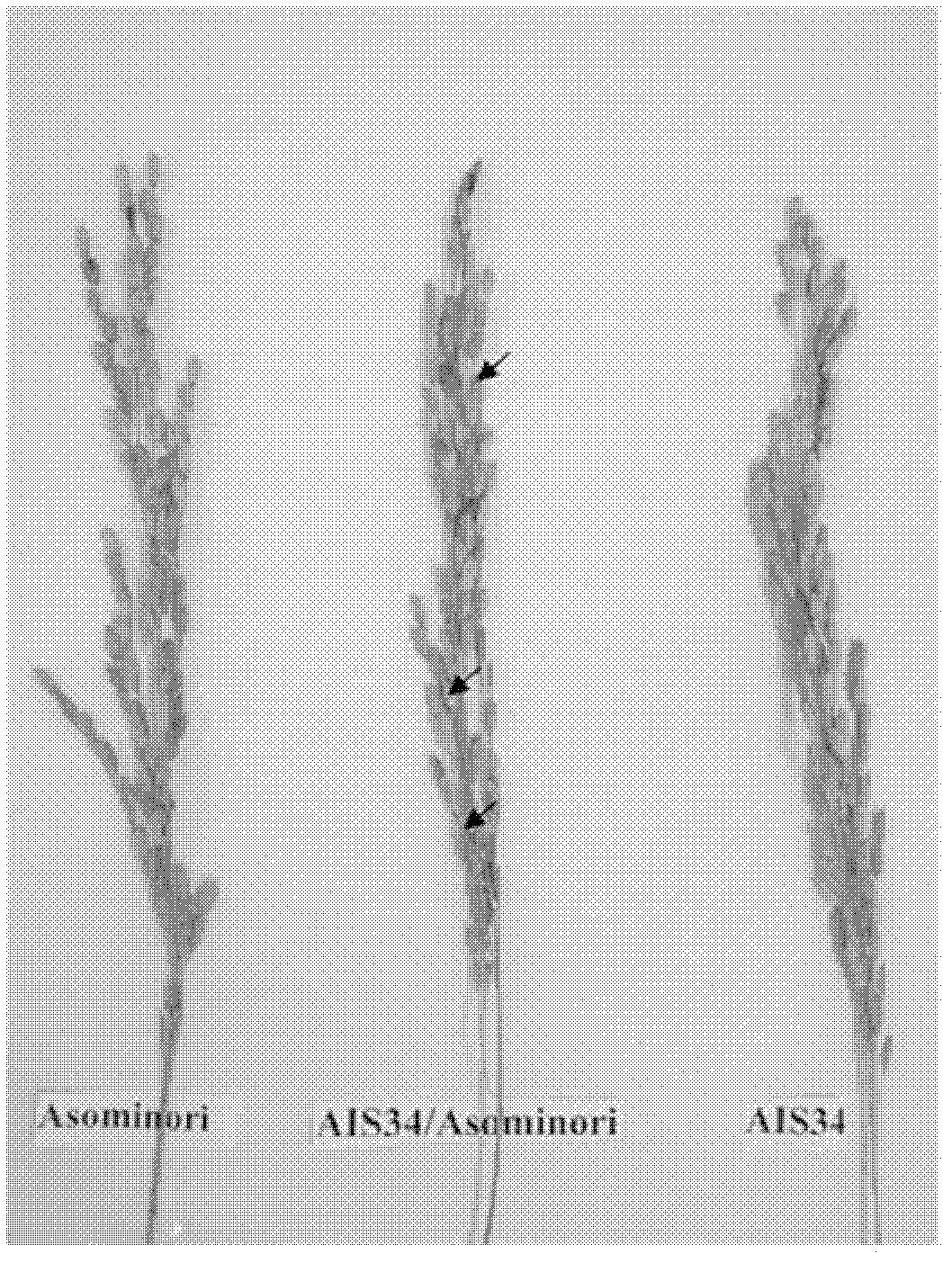
[0043] Using AIS34 to hybridize with Asominori, extract the leaf genome DNA of AIS34, Asominori and AIS34 / Asominori F1, according to the published SSR molecular marker linkage map and the published rice genome sequence, compared with the original map published by Kubo et al. (1999, 2002) , synthesize the corresponding primers on chromosome 5 (Table 1), for the two parents and F 1 For molecular marker detection, when multiple markers in a row are polymorphic between AIS34 and Asominori, and these markers are polymorphic in AIS34 / Asominori F 1 When the DNA band in is the same as that of Asominori, it can be considered that the replaced IR24 chromosome segment ends.
[0044] Table 1 Molecular markers for fine mapping
[0045]
[0046] Preliminary Mapping of AIS34 / Asominori Hybrid Sterility Gene
[0047] With AIS34 / Asominori / / Asominori backcross population, the AIS34 / Asominori hybrid F 1 Preliminary mapping of sterility genes.
[0048] Construction of Near-Isogenic Lines of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com