Improved penicillin antibiotic aptamer without fixed point target substance and application thereof
A technology of penicillin and antibiotics, applied in the direction of DNA/RNA fragments, recombinant DNA technology, color/spectral characteristic measurement, etc., can solve the problems of ssDNA loss and introduction of other substances, and achieve convenient operation, strong applicability, and good fixation effect Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
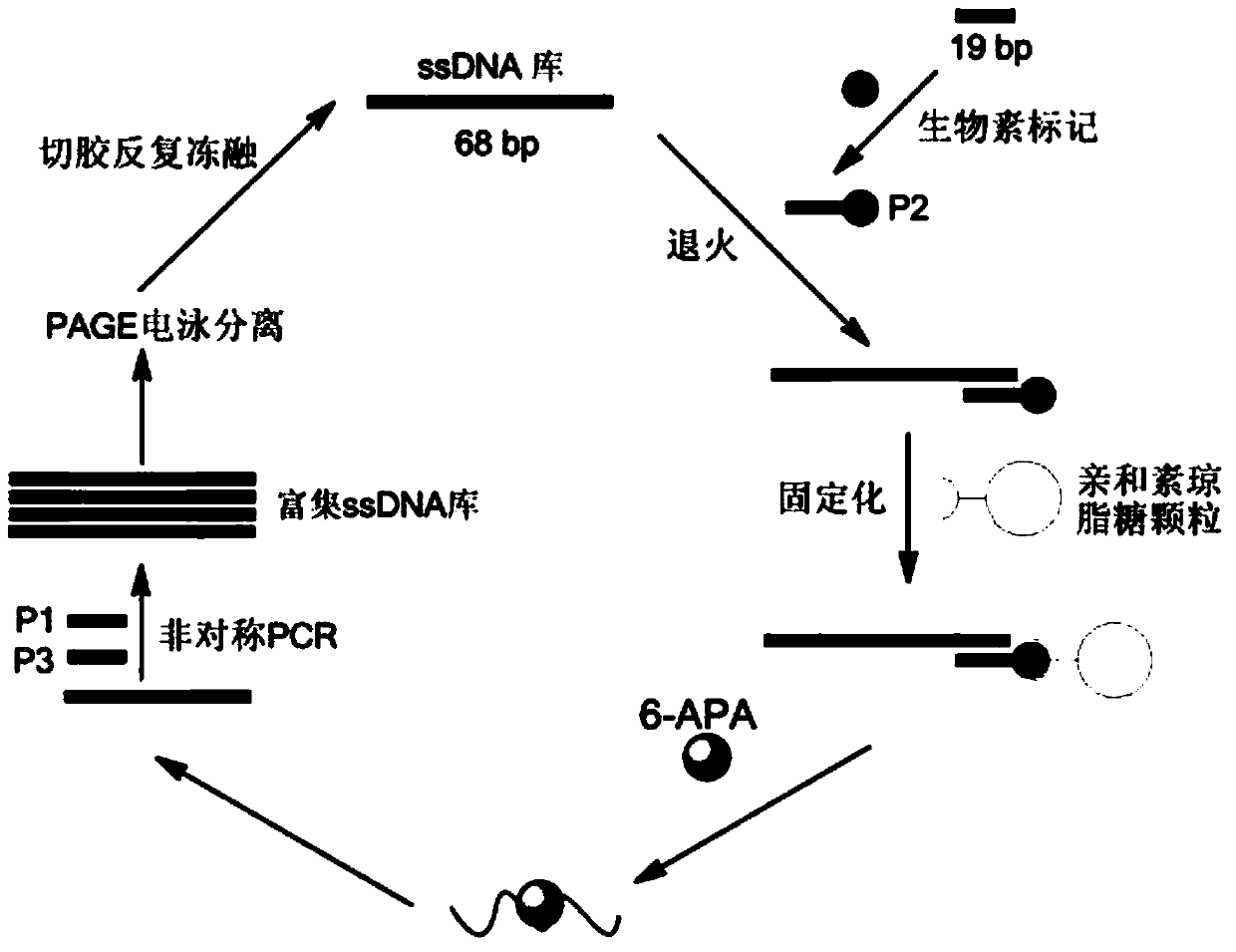
[0032] like figure 1 As shown, this embodiment includes the following steps:
[0033] 1) Construct a random oligonucleotide library with a total length of 68bp. The sequence of the oligonucleotide library is: 5'-ACCGACCGTGCTGGACTCT-N30-AGTATGAGCGAGCGTTGCG-3', wherein both ends are fixed sequences of 19bp, and the middle 30 bases is a random sequence.
[0034] 2) Design three primers, wherein: P1 (5'-ACCGACCGTGCTGGACTCT-3') and P2 (5'-CGCAACGCTCGCTCATACT-3') are used as the upstream primer and downstream primer of PCR amplification oligonucleotide respectively, and the upstream primer (5 '-Biotin-CGCAACGCTCGCT CATACT-3') was used to immobilize the oligonucleotide library on streptavidin-labeled agarose particles.
[0035] 3) Take a certain amount of random oligonucleotide library, and mix it evenly with the fixed sequence of agarose particles in a certain ratio (1:2~3), and anneal the mixture. The annealing conditions are: 94°C for 60 seconds, 59°C Anneal for 60 minutes, ext...
Embodiment 2
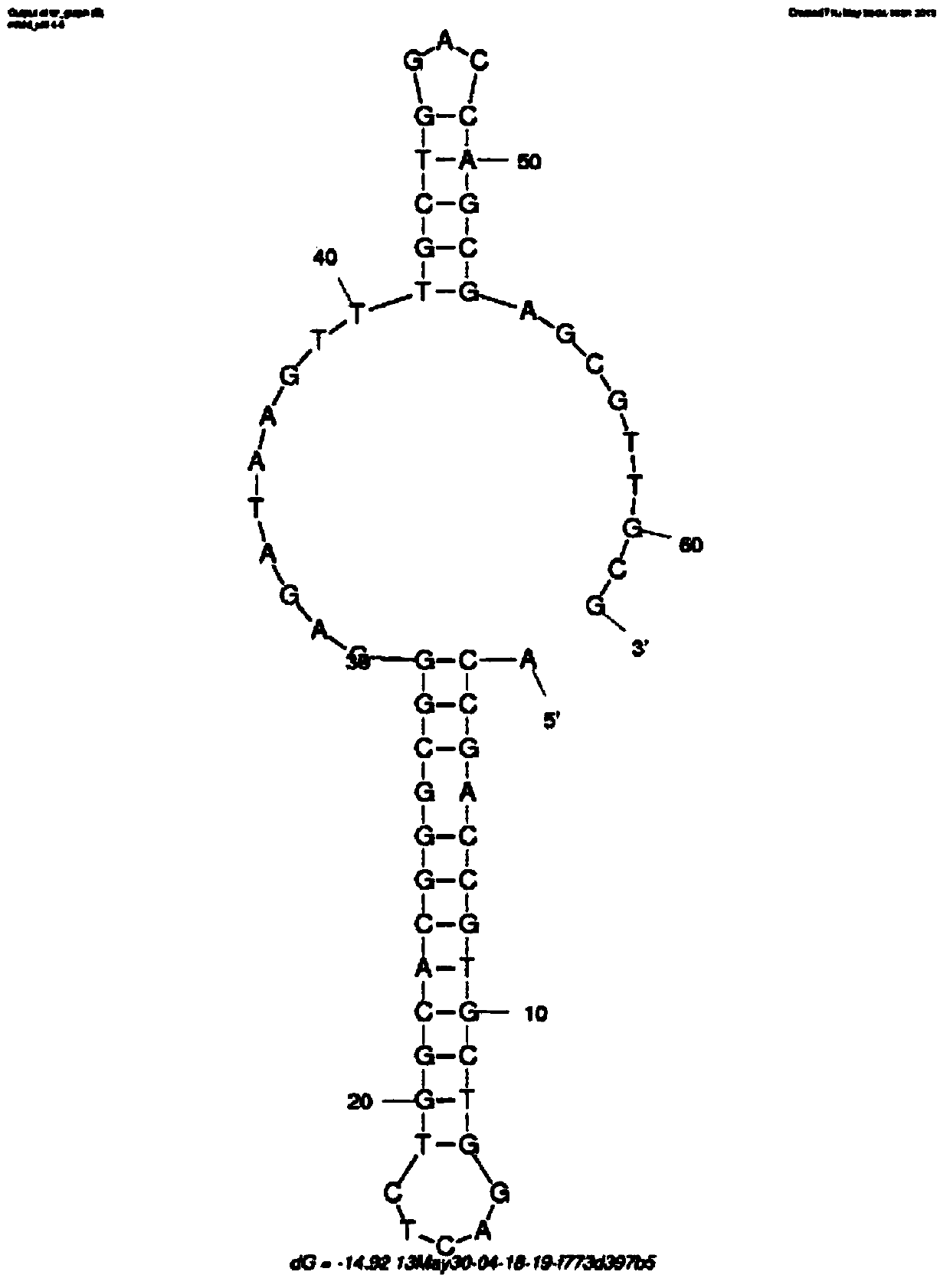
[0045] like figure 2 Shown is the specific description of the aptamer prepared in Example 1.
[0046] In this example, through cloning and sequencing, the base composition of the prepared aptamer is analyzed as shown in Seq ID No.1, specifically:
[0047] 5'-ACCGACCGTGCTGGACTCTGGCACGGGCGGGAGATAAGTTTGCTGGACCAGTATGAGCGAGCGTTGCG-3'
[0048] In this example, Mfold software was used to simulate the secondary structure of penicillin aptamers. And by calculation, the free energy of the penicillin antibiotic aptamer is -14.92, and the four bases A, T, C, and G are relatively evenly distributed. The number of T+G accounts for 43.6% of the total bases.
Embodiment 3
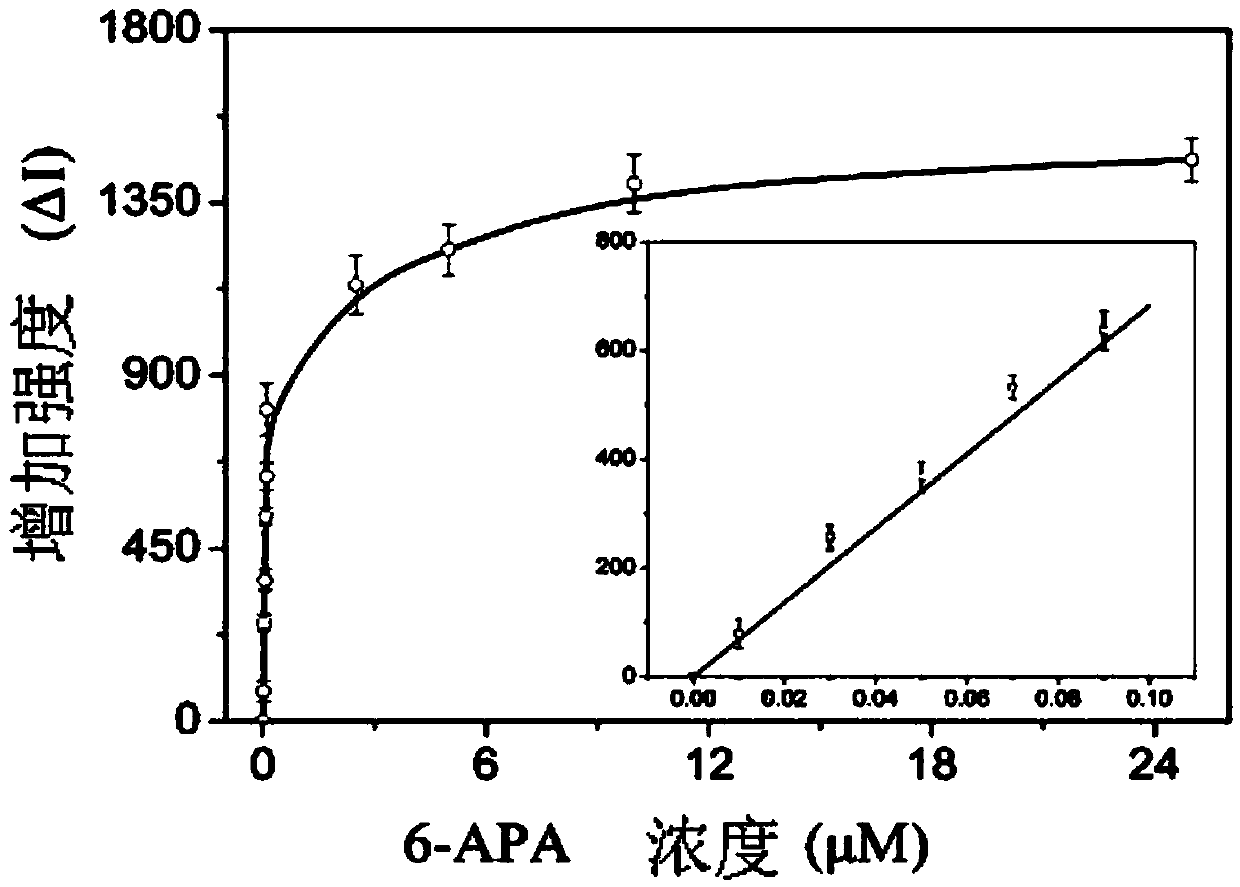
[0050] like figure 2 As shown, the specific application of the aptamer prepared in Example 1, that is, the penicillin aptamer sensor, its structure, composition and preparation method are
[0051] Optimizing the concentration of aptamers in the system: add penicillin antibiotic aptamers in the concentration range of 1-20nM to the nanogold solution, and finally add 0.1365μM PDDA, and use a microplate reader to measure the ratio of light absorption at 650nm and 520nm The curve was drawn for the difference, and the peak corresponding to the penicillin antibiotic aptamer concentration was 6nM.
[0052] Optimizing the PDDA concentration in the system: After incubating 6nM penicillin antibiotic aptamer with 0.2 and 2μM 6-APA solution for 15 minutes, add 100μL nano-gold, and finally add PDDA with a concentration range of 0.084-0.158nM for 5 minutes, then use The difference between the ratio of light absorption values at 650nm and 520nm measured by the microplate reader was drawn ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com