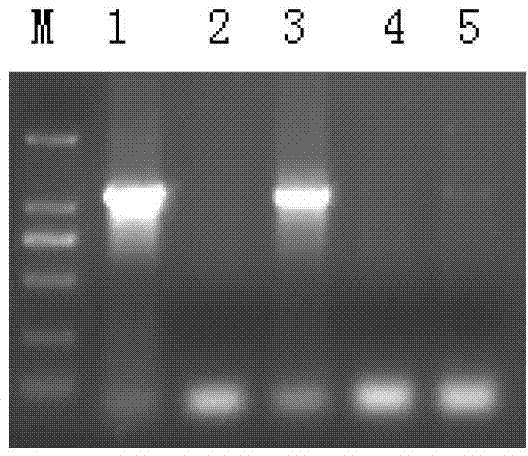
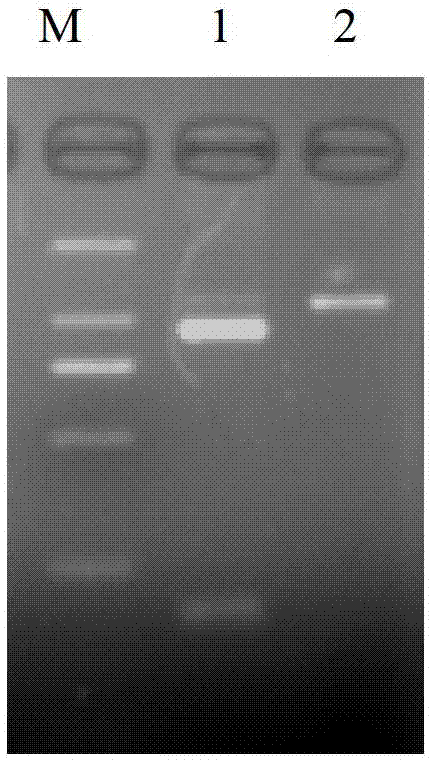
Identification method of channel catfish species
A technology for channel catfish and identification methods, which is applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of cumbersome operation process, lack of mitochondrial DNA molecular data, unsuitable for fast and economical identification of channel catfish species, etc. , to achieve the effect of short time and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0042] The principles and features of the present invention will be described below with reference to the accompanying drawings. The examples cited are only used to explain the present invention, and are not used to limit the scope of the present invention.
[0043] 1. Extraction of genomic DNA
[0044] The conventional phenol-chloroform extraction method is adopted. For specific reference, the method of Sambrook (2002) is slightly modified:
[0045] (1) Take about 0.1g of the tail fin fixed with alcohol in a 2ml centrifuge tube, cut into pieces, and dry at 37°C;
[0046] (2) Add 500ul extraction buffer to the centrifuge tube, and add RNaseA to a final concentration of 20μg / ml, and water bath at 37°C for 1h. Then add proteinase K to the extraction buffer to a final concentration of 200μg / ml, and bath at 55°C overnight;
[0047] (3) After cooling the digested sample to room temperature, add an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), gently invert it upside down for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com