Pair of specific primers and probe for detection of CYP2C19 gene chip
A CYP2C19, specific technology, applied in the field of molecular biology, can solve the problems of inaccurate detection results, long detection cycle, and difficulty in meeting clinical tests.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1 Preparation of gene chip
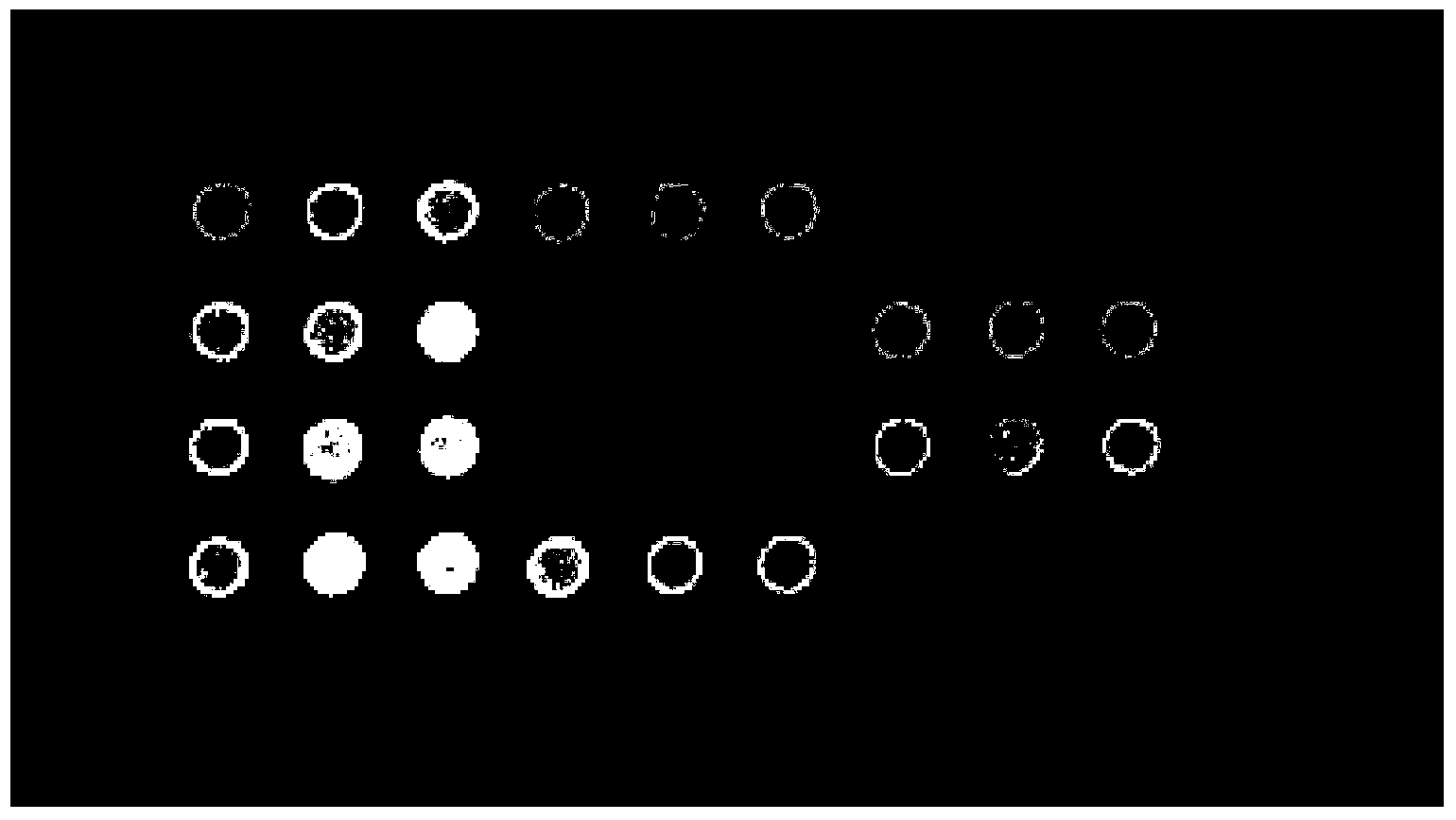
[0080] Aldehyde-modified glass slides (product number: BSM03011, Shanghai Bio Technology Co., Ltd.). The following probes were artificially synthesized (Shanghai Sangon Bioengineering Technology Service Co., Ltd.), dissolved in water to a concentration of 100pmol / ul, and then mixed in equal proportions with 2× spotting buffer (product number: BST02010, Shanghai Bio-Technology Co., Ltd.) . Then, use the GSM417 sample spotting instrument of Affymetrix Company to make points such as figure 1 array of . Leave overnight at room temperature.
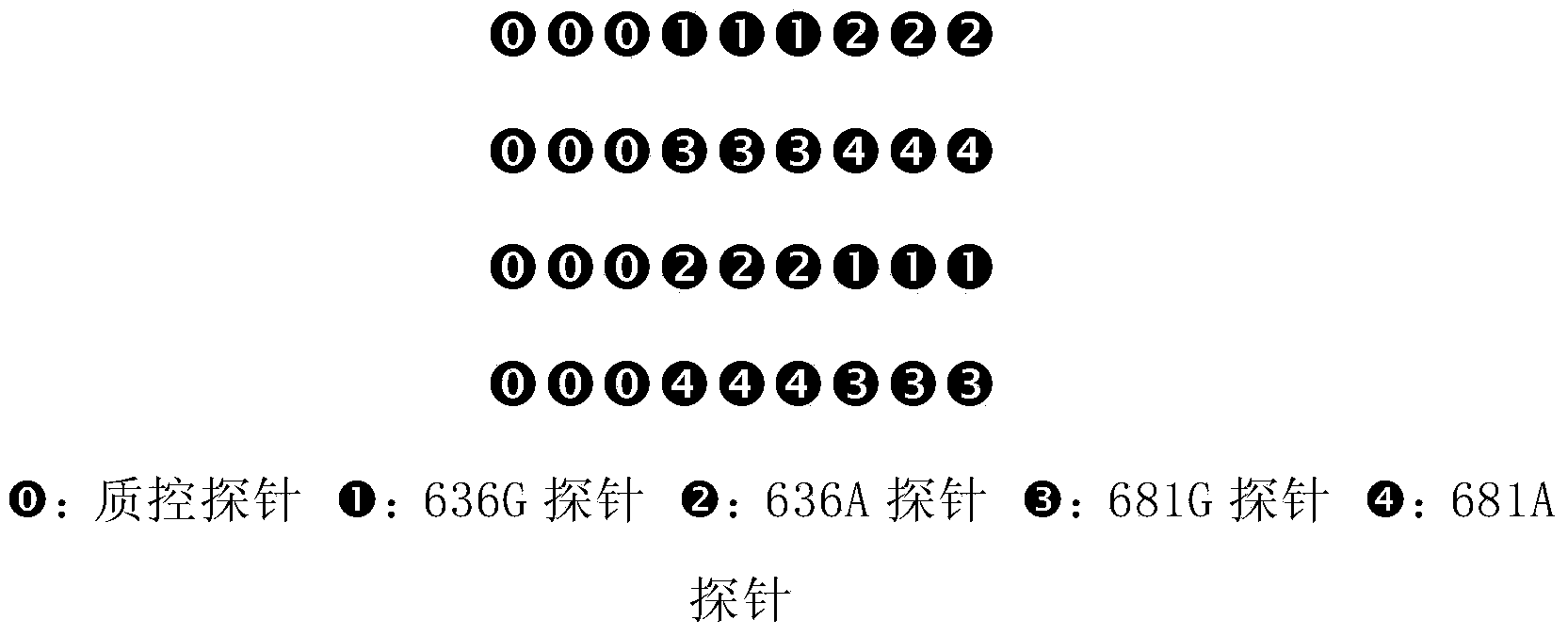
[0081] The sequences of each probe are as follows (the SNP sites are underlined).
[0082] The specific oligonucleotide probe for detecting the 681A site is: NH2-TTTTTTTTTTTTTTTTATTATTTCCC A GGAACCCAT;
[0083] The specific oligonucleotide probe for detecting the 681G site is: NH2-TTTTTTTTTTTTTTTTATTATTTCCC G GGAACCC;
[0084] The specific oligonucleotide probe for detecting the 636G site is:...
Embodiment 2
[0088] Example 2 Preparation of chromosomal DNA
[0089] Use the blood DNA extraction kit from Shanghai Bio-Tech Co., Ltd. and follow the instructions as follows:
[0090] Adsorption column activation:
[0091] Put the adsorption column in the collection tube, add 500 μL buffer BH1, let it stand for 2-3 minutes, and centrifuge at 12,000 rpm (9,500×g) for 30 s; discard the waste liquid in the collection tube, and put the adsorption column back into the collection tube. Add 500 μL of buffer BH2 to the adsorption column, centrifuge at 12,000 rpm (9,500×g) for 30 s, discard the waste liquid in the collection tube, and put the adsorption column back into the collection tube for use.
[0092] Operating procedure:
[0093] (1) Add 20 μL of proteinase K to the bottom of a 1.5 mL centrifuge tube with a pipette.
[0094] (2) Add 200 μL of blood sample into the centrifuge tube.
[0095] (3) Add 200 μL buffer BL to the centrifuge tube, shake and mix for 15 seconds.
[0096] (4) Inc...
Embodiment 3
[0105] Example 3 Using the primers provided by the present invention to amplify the CYP2C19 gene fragment by PCR method
[0106] Entrust Shanghai Sangon Bioengineering Technology Service Co., Ltd. to synthesize primers, and the primer information is as follows.
[0107] When used to detect the 681G / A site, the sequence of the primer pair used for amplification is:
[0108] Upstream SEQ ID No.13: 5'-CTTGGCATATTGTATCTATACCTTT-3'
[0109] Downstream SEQ ID No.14: 5'-CAAAACACAAATGATGCCTACAAAT-3'
[0110] When used to detect the 636G / A site, the sequence of the primer pair used for amplification is:
[0111] Upstream SEQ ID No.19: 5'-CACCCTGTGATCCCACTTTC-3'
[0112] Downstream SEQ ID No.20: 5'-TCTGTCGGTACCCCACTTAT-3'
[0113] And the 5' end of the primer pair was modified with biotin.
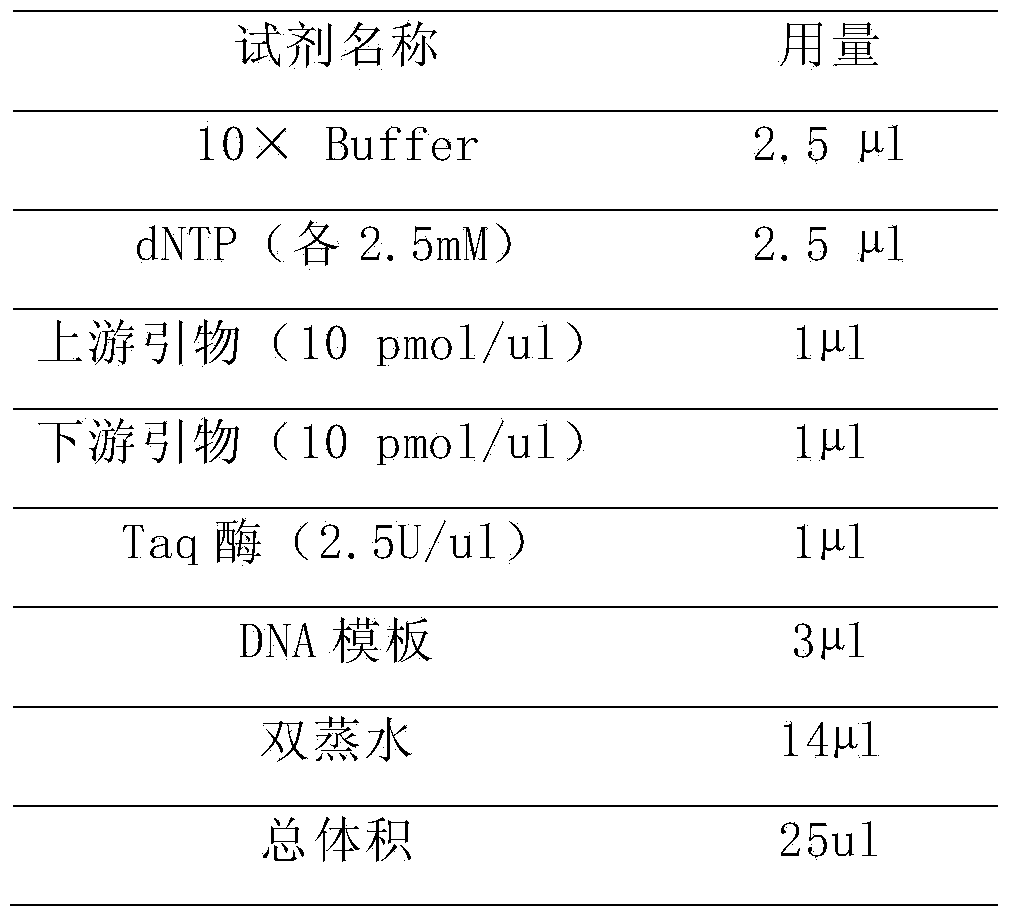
[0114] Then dissolve and dilute to 10 pmol / μl with water. Prepare the PCR amplification system with the purchased Taq enzyme (TaKaRa), 10× buffer (TaKaRa), dNTP (Shanghai Sangon Bioengineering...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com