Chemiluminescence reaction-based methylase detection probe, detection kit and detection method
A technology of methylase and chemiluminescence, which is applied in biochemical equipment and methods, measurement/inspection of microorganisms, DNA/RNA fragments, etc., can solve problems such as time-consuming, complicated pretreatment of gold nanoparticles, and unsafety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
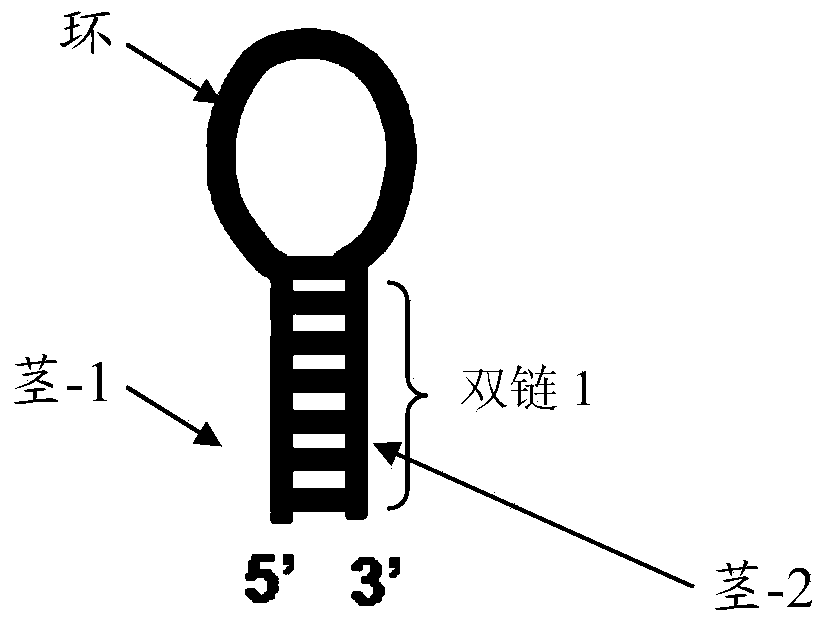
[0147] figure 1 Schematic diagram of the structure of the double-strand 1 formed by the 5' end side chain (stem-1) and the 3' end side chain (stem-2) of the hairpin probe provided in the embodiment of the present invention. The hairpin probe includes double-strand 1 and ring. Wherein, the double-stranded DNA1 has a methylase recognition site and a methylation-dependent endonuclease cleavage sequence, wherein the distance between the methylase recognition site and the terminal base of the loop is 2~8bp;
[0148] combine figure 1 Taking Dam methylase as an example, this embodiment provides a methylase detection probe based on chemiluminescence reaction, including the following steps:
[0149] (1) Synthesize the hairpin probe in vitro, the nucleotide sequence (5' end to 3') is shown in SEQ ID NO:12;
[0150] (2) Design primer-1 and primer-2 according to the sequence of the hairpin probe. As long as primer-1 includes the nicking enzyme cleavage site and the site complementary ...
Embodiment 2
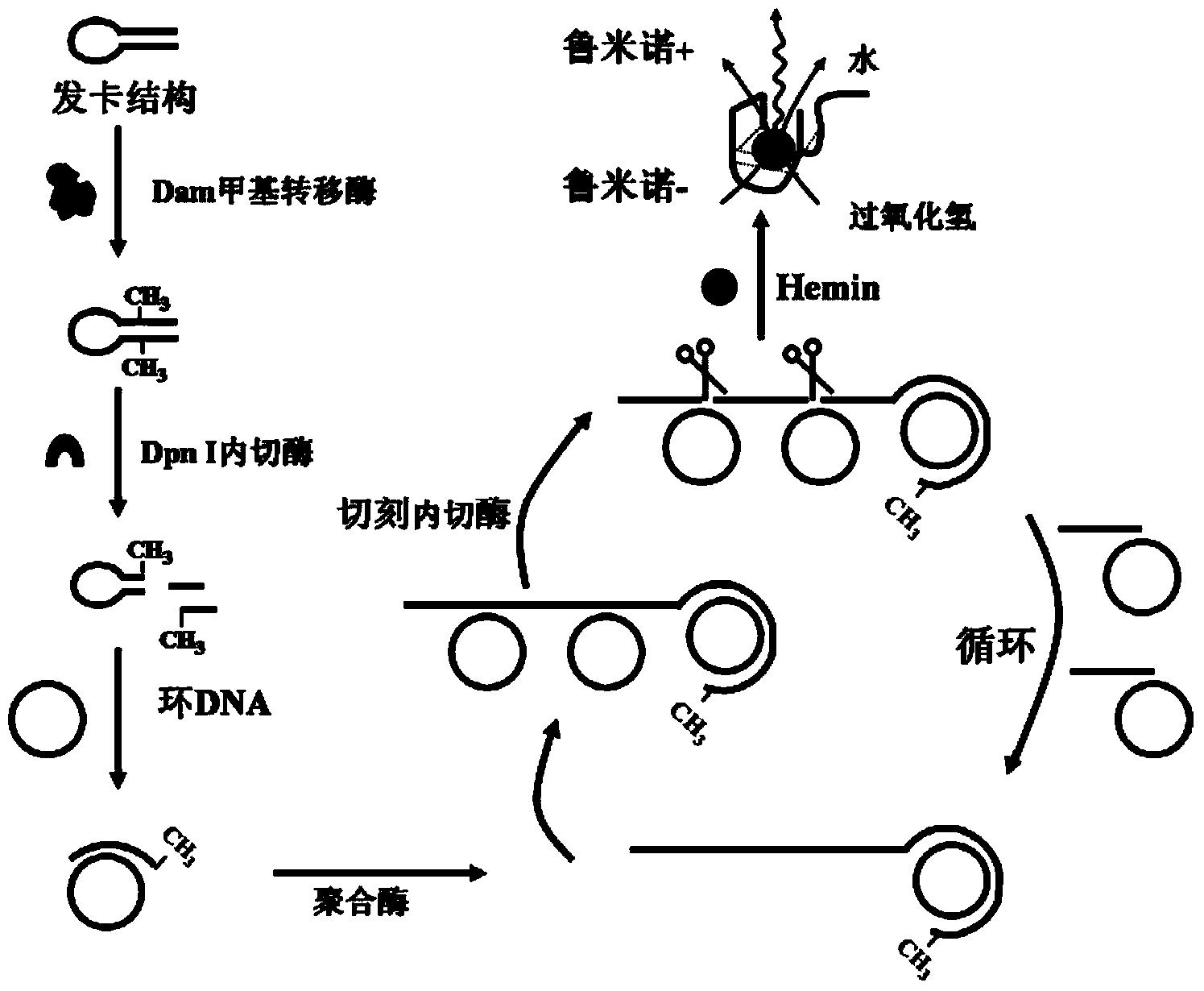
[0161] Taking Dam methylase as an example, a flow chart of a DNA peroxidase-based methylase detection method is provided, as shown in figure 2 shown. Depend on figure 2 It can be seen that this detection method can be roughly divided into three steps. The first step is methylation and methylation-sensitive endonuclease digestion reaction: the sequence GATC on the stem structure of the hairpin probe can be specifically identified by the methyltransferase Dam To recognize and add a methyl group to the double-stranded A base; the methylated hairpin structure can be recognized and cut by endonuclease Dpn Ⅰ, resulting in a small hairpin structure and double-stranded DNA, due to The annealing temperature of the small hairpin structure and double-stranded DNA is too low, causing it to transform into single-stranded DNA at room temperature. The second step is the rolling circle amplification reaction and the nicking enzyme digestion reaction: the single-stranded DNA transformed in...
Embodiment 3
[0178] This embodiment provides a method for detecting methylase based on DNA peroxidase, which can directly obtain the concentration of methylase by preparing a standard curve, including the following steps:
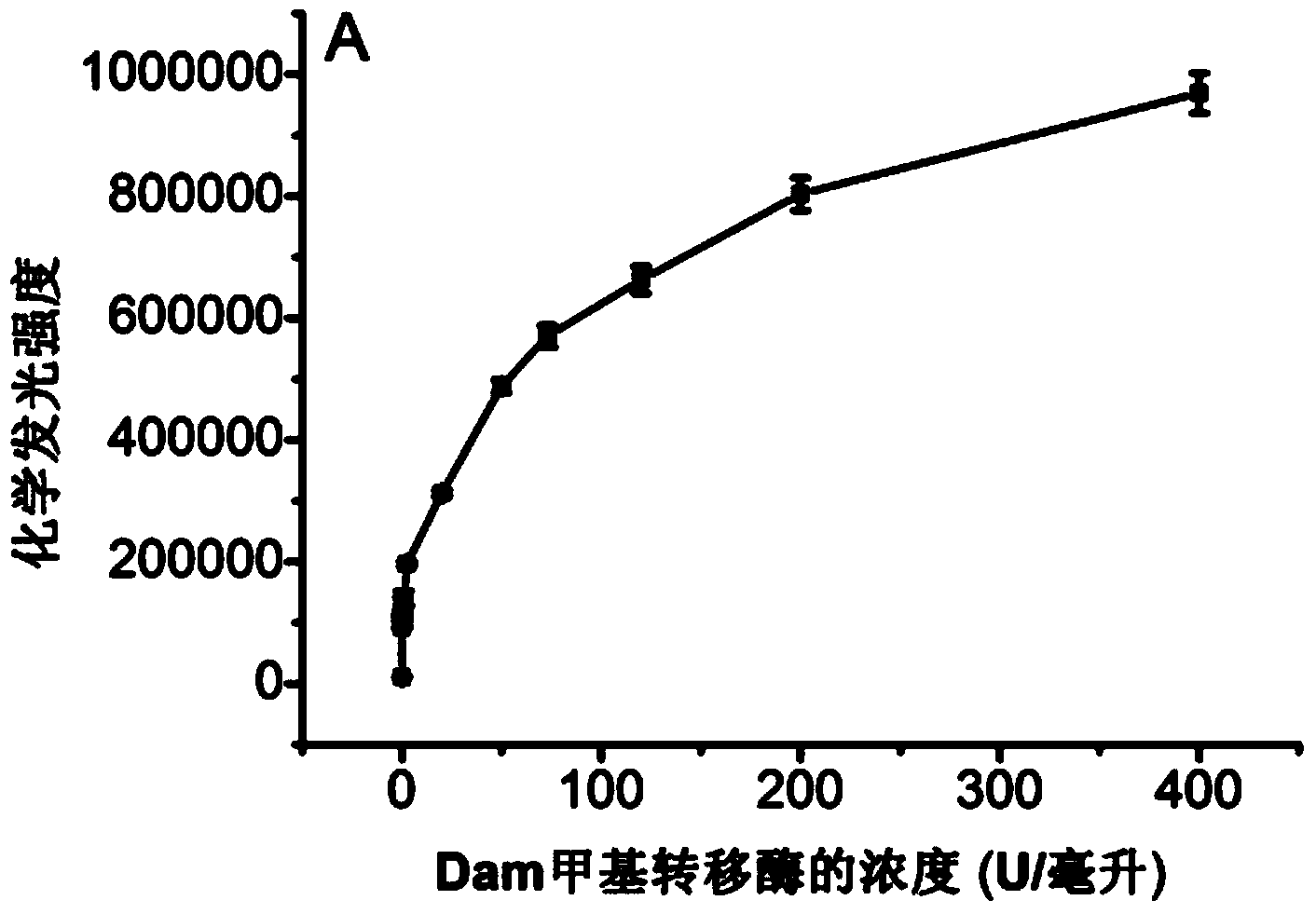
[0179] Prepare the methylase into a solution of known concentration (the concentration of methylase is 0.025, 0.05, 0.1, 0.5, 1, 2, 2.5, 25, 50, 73, 120, 120, 200, 400 units per milliliter), Using the DNA peroxidase-based methylase detection kit provided in Example 2 and the detection method using the kit to detect the chemiluminescence signals of the methylase solutions at various concentrations, such as image 3 shown, and prepare a standard curve, such as Figure 4 shown. image 3 The relationship between different concentrations of methylase and the intensity of chemiluminescence is shown (the error bars represent the standard deviation of three experiments), and the concentration range is from 0.025 to 0.5 units per milliliter. Figure 4 The logarithmic value of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com