Fluorescent sensor for detecting HBV, and preparation and applications thereof
A fluorescent sensor and fluorescent signal technology, applied in the field of HBV detection, can solve the problems of reaction efficiency, limiting the versatility of nucleic acid analysis, and unfavorable promotion.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Fabrication of fluorescent sensors and detection of miRNA
[0046] 1. Materials and methods
[0047] 1.1 Materials
[0048] HPLC-purified oligonucleotides were synthesized by Shanghai Sangong. EnGen Lba Cas12a and NEBuffer 3.0 solutions were purchased from New England Biolabs. DEPC water, enzyme-free water and RNase inhibitors were purchased from Shanghai Sangong.
[0049] 1.2 Testing instruments
[0050] Edinburgh Integrated Steady State Transient Fluorescence Spectrometer FS5.
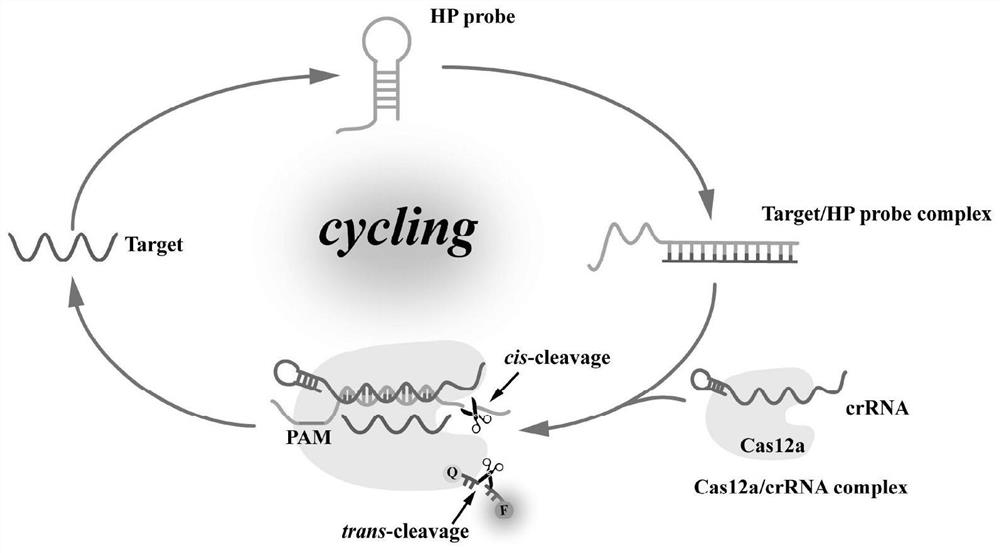
[0051] 1.3 Detection principle
[0052] Such as figure 1 As shown, in the homogeneous system, the target substance binds to the toehold in the HP probe, and the HP probe is opened by the toehold-mediated strand displacement reaction, thereby exposing the toeholds located in the HP probe that are closed. Subsequently, the preassembled Cas12a / crRNA hybridizes to the exposed toehold domain via crRNA, displacing the target with base-pairing extension and forming the Cas12a activator. Then,...
Embodiment 2
[0070] Validation of the feasibility of detecting miRNA fluorescent sensors
[0071] 1. The verification of foothold-mediated cascade strand displacement reaction and Cas12a cleavage, the process is as follows:
[0072] 1) Preparation of HP probe: NEBuffer 3.0 buffer was used to prepare HP into a 20 μL solution with a concentration of 10 μM, denatured at 95° C. for 5 minutes, slowly cooled to room temperature and placed at 4° C. for later use. 2) Fluorescence spectrophotometer to verify the feasibility of the experiment:
[0073]
[0074]
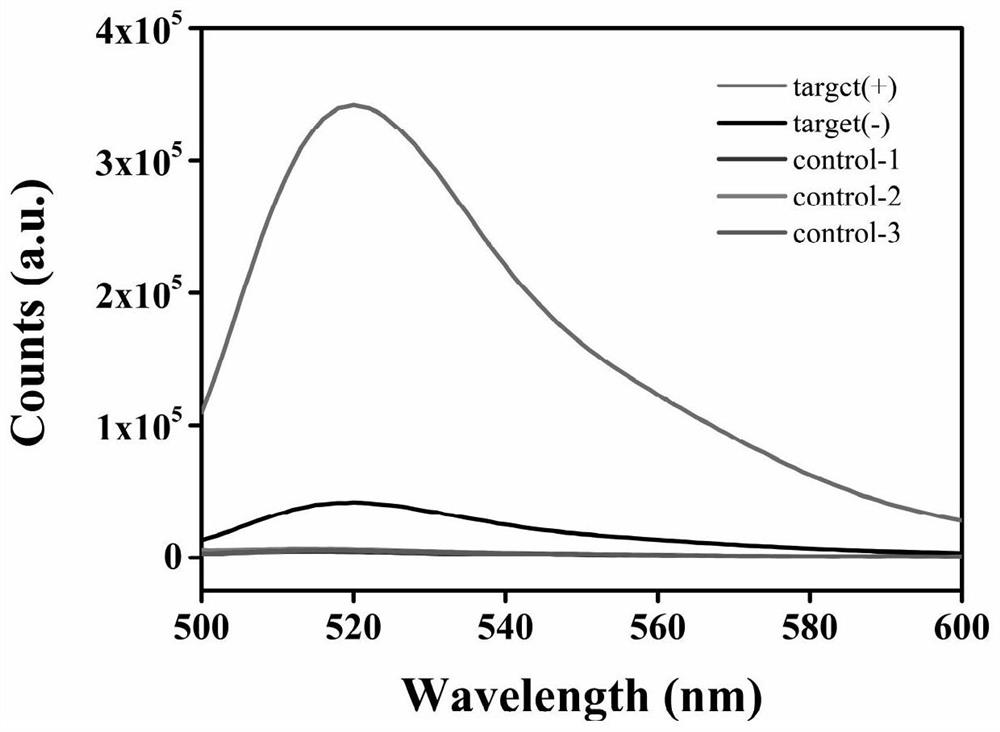
[0075] Add the above liquid accordingly to obtain 100 μL of working solution, mix it on a shaker, centrifuge it quickly with a hand-held centrifuge for a short time, and place it in a 37°C incubator for 1 hour of reaction. After the reaction was completed, the fluorescence intensity of the solution was measured using a fluorescence spectrophotometer. The result is as figure 2 shown. figure 2 Among them, the red curve is the expe...
Embodiment 3
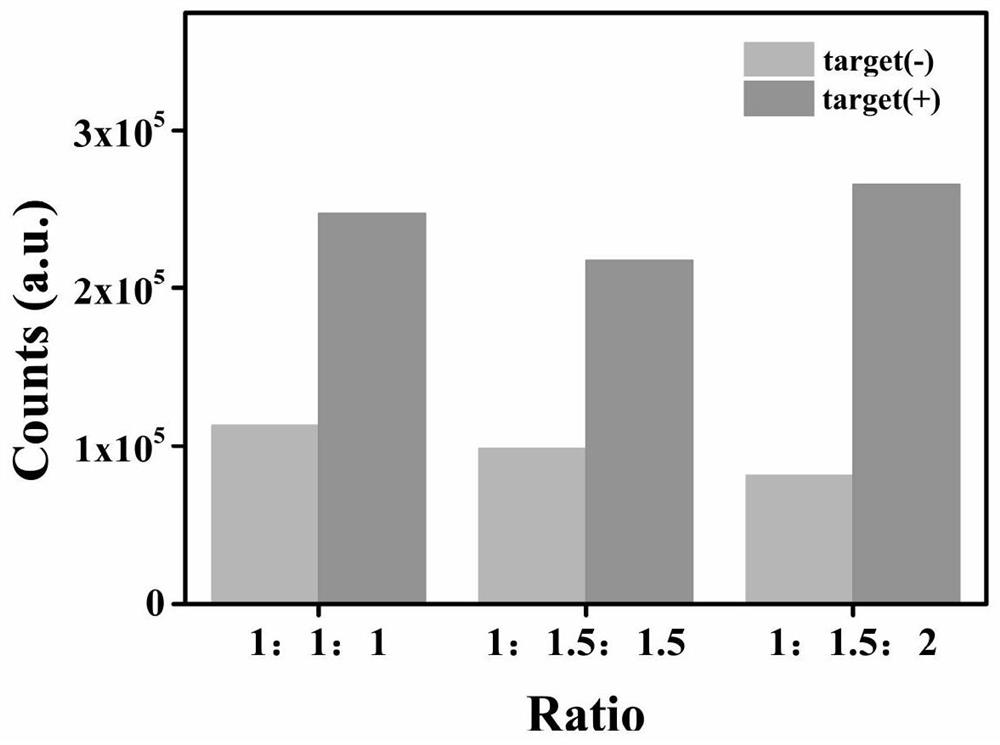
[0077] Molar ratio of HP probe, Cas12a, crRNA
[0078] In order to investigate the influence of the molar ratio of HP probe, Cas12a, and crRNA on the stability and reaction speed of the developed fluorescent sensor, this example further studies the optimal molar ratio of HP probe, Cas12a, and crRNA.
[0079] Depend on Figure 4 It can be seen that when the molar ratio is 1:1.5:2, the signal-to-noise ratio is the best. Therefore, a molar ratio of 1:1.5:2 was selected as the optimum ratio.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com