Site-directed mutagenesis system of mustard genome and application of site-directed mutagenesis system
A genome-specific, gene-based technology, which is applied in genetic engineering, recombinant DNA technology, and the introduction of foreign genetic material using vectors, can solve the problem of mustard gene editing and transformation technology that has not yet been reported, and achieve the effect of enriching genetic resources.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Embodiment 1, the acquisition of mustard FRIGIDA gene fragment
[0017] A fragment of the mustard FRIGIDA-b gene containing the first exon and the first intron was amplified by a primer pair (upstream 5'‐TGCCTACAAACACGGAAAT‐3', downstream 5'‐AAGGGCCATACAAATGCTAT‐3'). This example takes the first exon as the target point.
Embodiment 2
[0018] Embodiment 2, the design of the first exon nuclease TALEN-L and TALEN-R
[0019] According to the characteristics of the first exon sequence of the mustard FRIGIDA gene, the 3' end sequence of the first exon was selected as the target sequence of the transcription activator effector nuclease TALEN-L and TALEN-R. According to the TALEN recognition principle, the designed TALEN recognition target sequence is 5'‐TGAGATTGCTGCTGCTctaaaacggtcacctttccttgtCCCTATGATGTCAGGTA‐3', the uppercase part is the recognition module of the transcription activator effector nuclease TALEN‐L and TALEN‐R, and the lowercase part is the spacer sequence, where the 5' The TALEN-L recognition module (SEQ ID NO1) at the end, and the TALEN-R recognition module (SEQ ID NO4) at the 3' end. Finally, the nuclear localization signal was set upstream of the TALEN protein, and the nuclease FokI was set downstream to obtain TALEN-L and TALEN-R with recognition modules and fusion nuclease FokI. Design the co...
Embodiment 3
[0023] Example 3, TALEN-L and TALEN-R functional verification
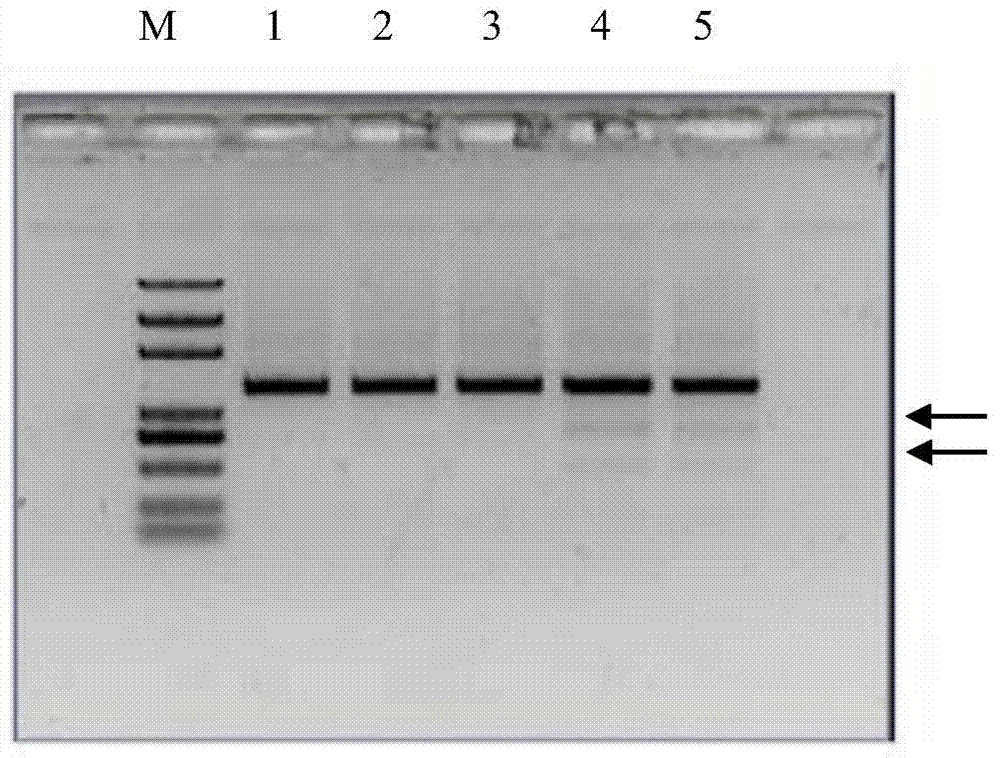
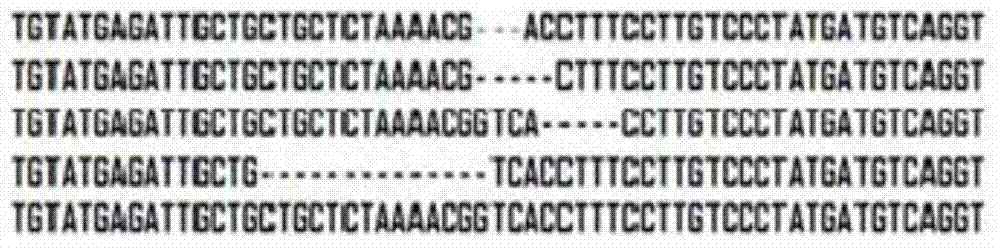
[0024] The binary expression vectors of TALEN-L and TALEN-R were constructed to transform Agrobacterium GV3101, and the positive recombinants were detected to infect the hypocotyls of Brassica juncea. Induce the regeneration plant to extract the genome, use T7 endonuclease to detect mutations, the detection system is T7E1buffer2μl, DNA2μl, T7E11μl, ddH 2 05 μl. Then use the primer pair (upstream 5'‐TGCCTACAAACACGGAAAT‐3', downstream 5'‐AAGGGCCATACAAATGCTAT‐3') to amplify the mutant FRIGIDA gene fragment, the PCR reaction system is: ddH 2 O32.8μl, 10×PCR buffer5μl, 2mM dNTP5μl, 25mM MgSO43μl, upstream and downstream primers 1.5μl, KOD DNAPolymerase (Toyobo)1μl, template DNA1μl. The fragment was then cloned into the pMD19‐T vector for sequencing, and mutants (such as figure 1 , figure 2 shown).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com