Application of marine-derived oligoguluronic acid
A technology of guluronic acid and source, applied in the field of biomedicine, can solve problems such as less reports on oligomer activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0020] Preparation of oligomeric guluronic acid from marine sources
[0021] Sodium alginate (sodium alginate) is a water-soluble polysaccharide derived from the cell wall of brown algae. It is a linear acidic polysaccharide formed by connecting D-mannuronic acid and L-guluronic acid through 1→4 glycosidic bonds. The marine-derived oligoguluronic acid (marine-derived oligoguluronic acid, MdOA) of the present invention is obtained from brown algae-derived sodium alginate through H 2 o 2 Cracked, the structure is as follows:
[0022]
Embodiment 1
[0023] Example 1 Detection of NO generation in cells by Griess method
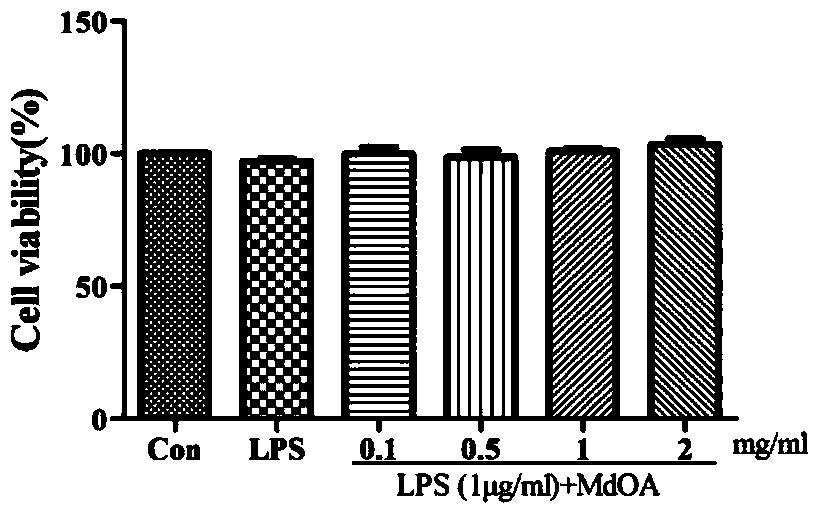
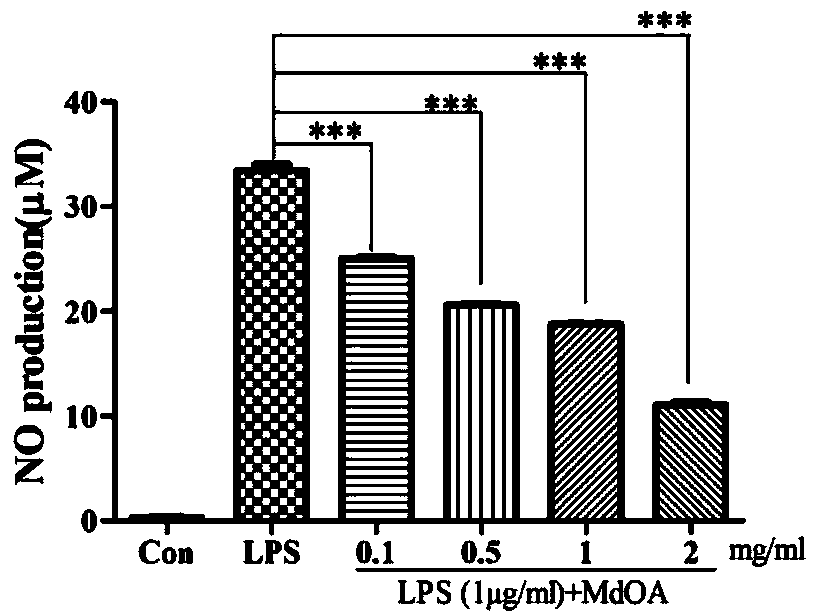
[0024] RAW264.7 cells (2×10 5 per well) adhered to the wall in a 96-well plate, discarded the supernatant after 4-6h, added 1mg / ml of MdOA, used serum-free RPMI1640 medium as the negative control group (Con), and added only 1μg / ml LPS as the negative control group (Con). Positive control group (LPS), with the addition of 1 μg / ml LPS and different concentrations of MdOA (0.1mg / ml, 0.5mg / ml, 1.0mg / ml, 2.0mg / ml) as the experimental group (LPS (1μg / ml) + MdOA), after stimulation for 24h, the production of NO in the medium was detected by the Griess method.
[0025] Such as Figure 1A As shown, the CCK-8 cell viability detection kit showed that compared with the Con group, LPS and different concentrations of MdOA had no toxicity to the cells.
[0026] In the concentration gradient experiment, different concentrations of MdOA (0.1mg / ml, 0.5mg / ml, 1.0mg / ml, 2.0mg / ml) stimulated RAW264.7 cells for 24 hours, and ...
Embodiment 2
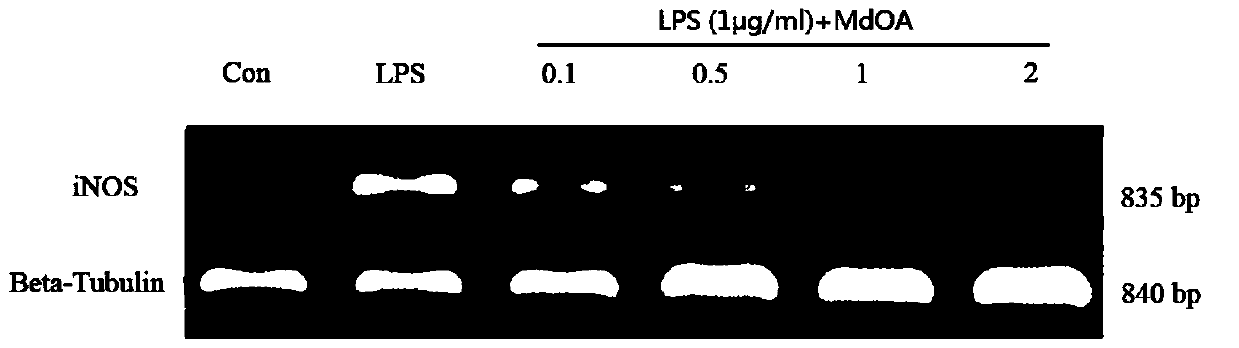
[0027] Example 2 Detection of the production of inflammation-related genes in cells by reverse transcription method (RT-PCR)
[0028] RAW264.7 cells (2×10 6 Each well) attached to the wall in a 6-well plate, after 4-6 hours, discard the supernatant, add different concentrations of MdOA, use serum-free RPMI1640 medium as the negative control group (Con), and add only 1 μg / ml LPS as positive For the control group (LPS), after being stimulated for 12 hours, total RNA was extracted according to the total RNA cell rapid extraction kit. Then, the total RNA was reverse-transcribed into cDNA by a reverse transcription kit, and then the genes of iNOS and COX-2 were obtained by PCR.
[0029] iNOS primers are: FE: 5'-CAACCAGTATTATGGCTCCT-3',
[0030] RE: 5'-GTGACAGCCCGGTCTTTCCA-3';
[0031] Primer for β-actin: FE: 5'-GGAGAAGATCTGGCACCACACC-3';
[0032] RE: 5'-CCTGCTTGCTGATCCACATCTGCTGG-3');
[0033] COX-2 primer: FE5'-CCACTTCAAGGGAGTCTGGA-3';
[0034] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com