Kit used for detecting number of human chromosomes 21
A technology for the number of chromosomes and detection reagents, applied in the field of molecular biology, can solve the problems of DNA quality difference, inaccuracy of results, annealing temperature changes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Embodiment 1: Using the kit of the present invention to detect normal specimens and specimens to be tested for trisomy 21
[0020] 1. The composition of the kit:
[0021] 1.1 Selection of target sequence and design of primers and probes
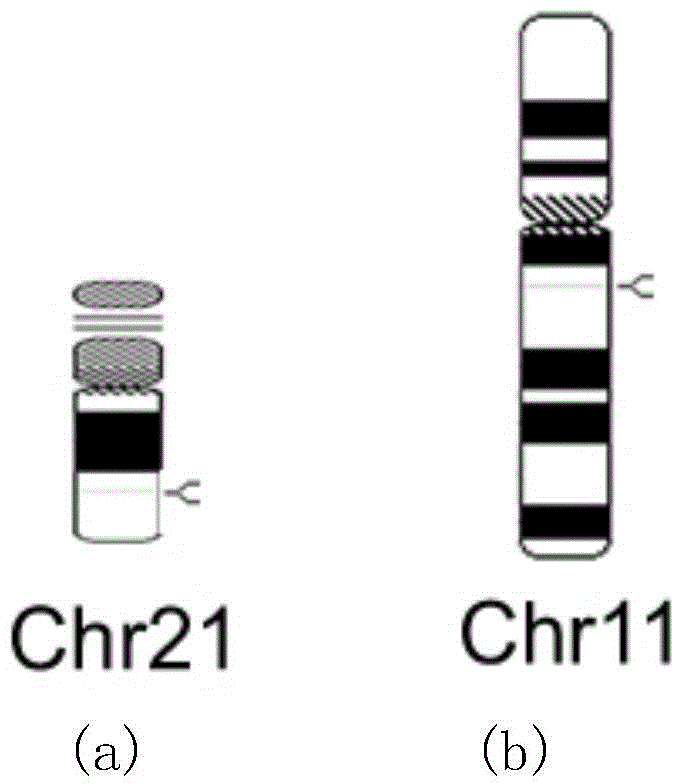
[0022] Detection sites: select the specific repetitive sequence chr21-2 on chromosome 21 and the specific repetitive sequence chr11-1 on chromosome 11 (chr21-2 and chr11-1 are two similar sequences) for relative real-time fluorescent quantitative PCR amplification and detection sequence, such as figure 1 shown, where:
[0023] The base sequence of the specific repetitive sequence chr21-2 on chromosome 21 is: 5'-gtgccattgacacaggaggacccatgcctgagaaagacttttatctgagttatactgagattaagatgttttgaagctcccaagcagagggatgctggatctgctgtggaaatctggctgagcagctgcagggacagctgggggtaaag-3' (SEQ ID NO: 5);
[0024] The base sequence of the specific repetitive sequence chr11-1 on chromosome 11 is: 5'-gtgccattgacacaggaggacctacgcctcagaaagacttttatctgagttaccaaggttgag...
Embodiment 2
[0046] Example 2: Using the kit, method, steps, etc. described in Example 1 to detect clinical samples
[0047] The kit was used to test 35 normal specimens and 26 trisomy 21 specimens, all of which were derived from DNA samples whose karyotypes were determined by traditional karyotype analysis methods.
[0048] Relative Quantification ΔCT by Chromosome 21 vs. Chromosome 11 21-11 Value (△CT 21-11 =Ct ROX -Ct HEX ), it can be judged whether the specimen is a normal specimen or a 21-trisomy specimen. The real-time fluorescence quantitative detection △CT value and its range are as follows:
[0049] △CT of chromosome 21 and chromosome 11 in normal specimen 21-11 The value ranges from -0.98 to -0.70; the △CT of chromosome 21 and chromosome 11 in trisomy 21 specimens 21-11 The value range is -1.71~-1.39, and the △CT value between the normal specimen and the 21-trisomy specimen has no overlap, and the distribution of the △CT value in the interval range can be used to judge wheth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com