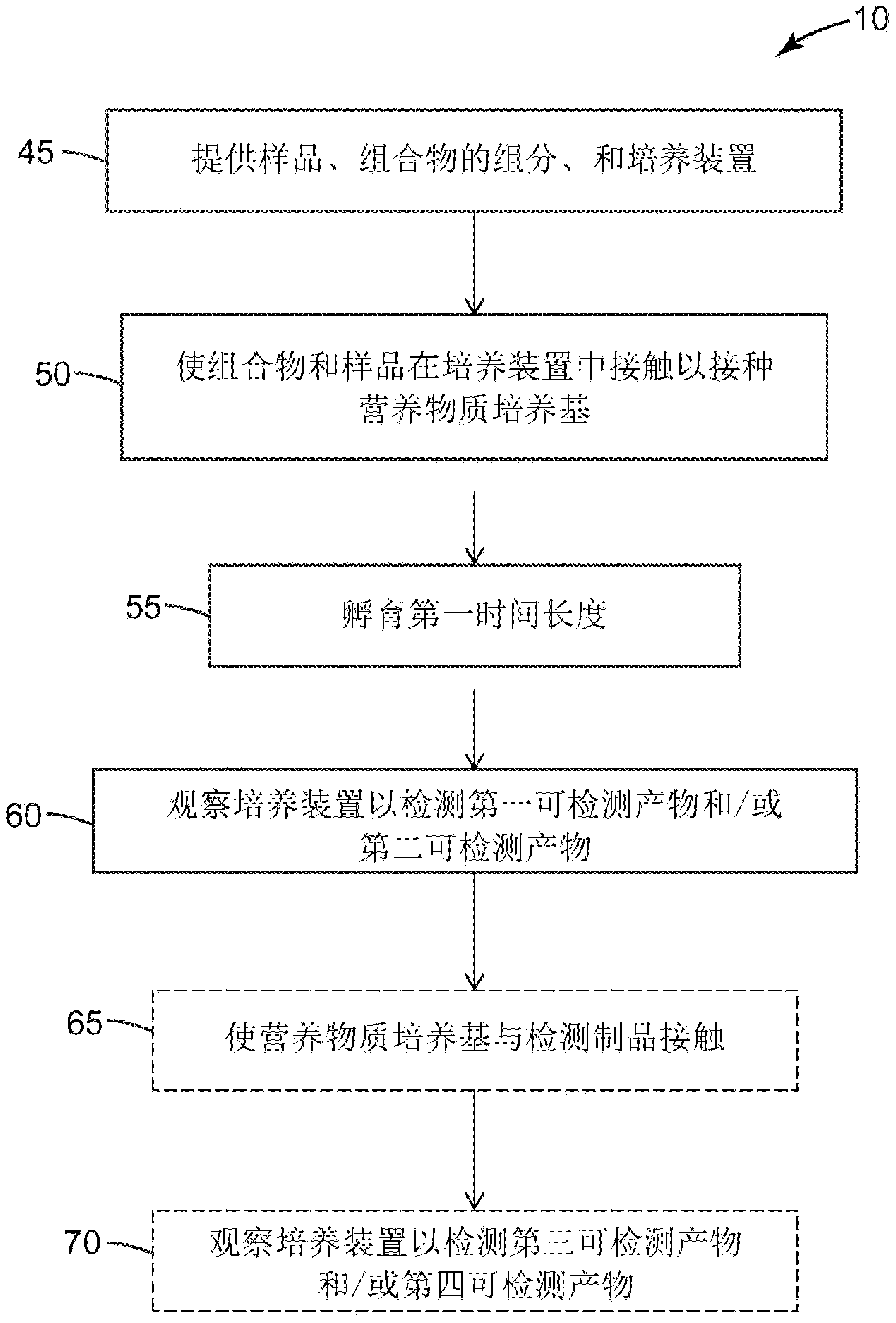
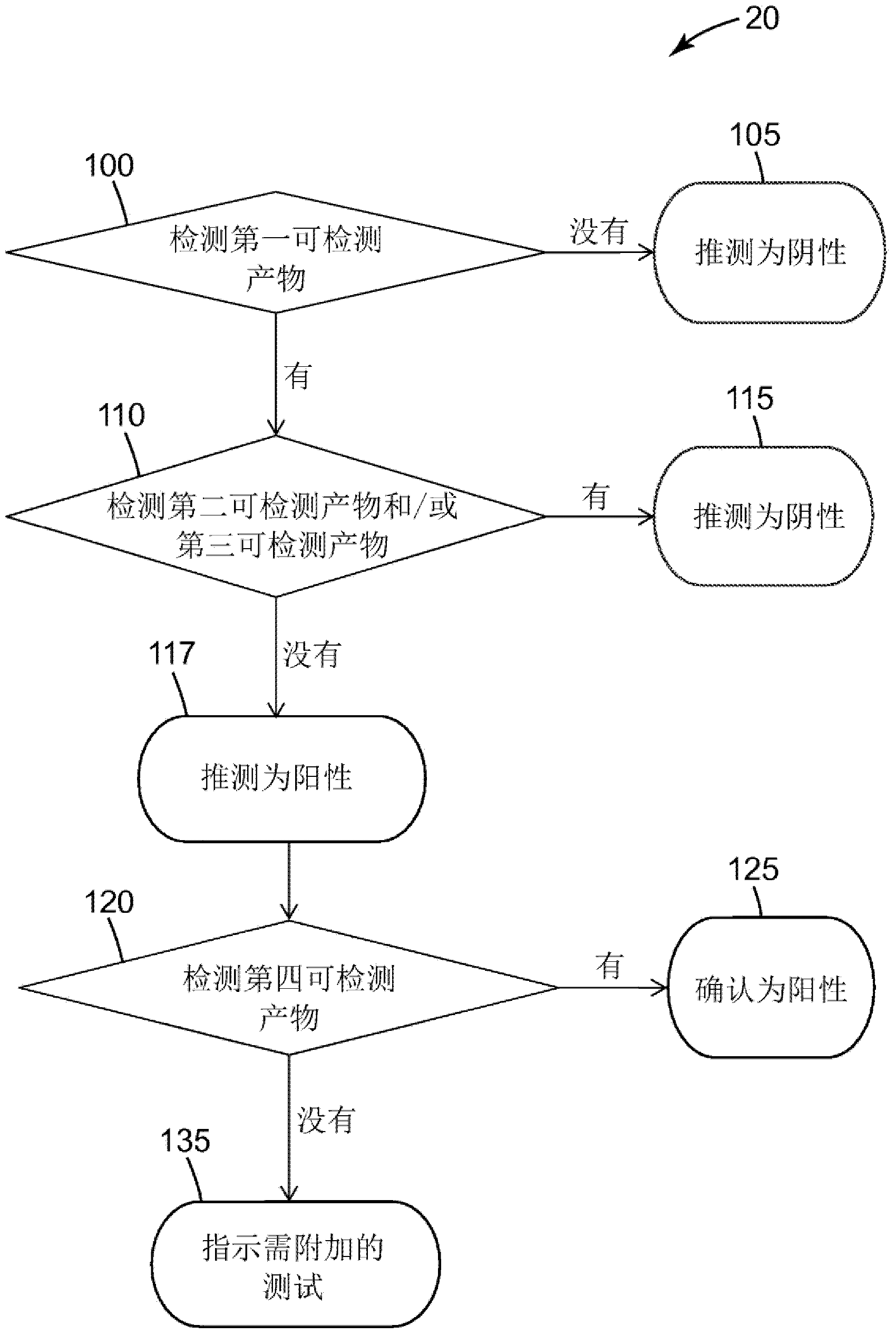
Method of detecting a salmonella microorganism
A technology for Salmonella and microorganisms, which is applied in the field of compositions with or without existence, and can solve the problems such as the difficulty in detecting Salmonella microorganisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1 is a composition comprising:
[0086] semi-solid medium comprising:
[0087] gelling agent;
[0088] at least one first selection agent that inhibits the growth of Gram-positive microorganisms;
[0089] a first differential indicator system comprising at least one first differential indicator compound; and
[0090] a second differential indicator system comprising a second differential indicator compound that is converted by urease activity to a second detectable product;
[0091] wherein said first differential indicator compound is capable of being converted by members of the genus Salmonella into a first detectable product;
[0092] wherein said first differential indicator compound is not capable of being transformed by genera of non-Salmonella, Gram-negative enteric microorganisms that form detectable colonies in and / or on said culture medium.
Embodiment 2
[0093] Embodiment 2 is the composition of embodiment 1, wherein the first differential indicator compound is selected from the group consisting of melibiose, 2-deoxy-D-ribose, mannitol, L-arabinose, galactose Alcohol, maltose, L-rhamnose, trehalose, D-xylose, sorbitol, and any combination of two or more of the above compounds.
Embodiment 3
[0094] Embodiment 3 is the composition of embodiment 1 or embodiment 2, further comprising a buffer.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com