DNase high-throughput sequencing detection signal processing method of DNA protein binding sites
A protein binding site, high-pass sequencing technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, informatics, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] The present invention will be described in further detail below in conjunction with the accompanying drawings.
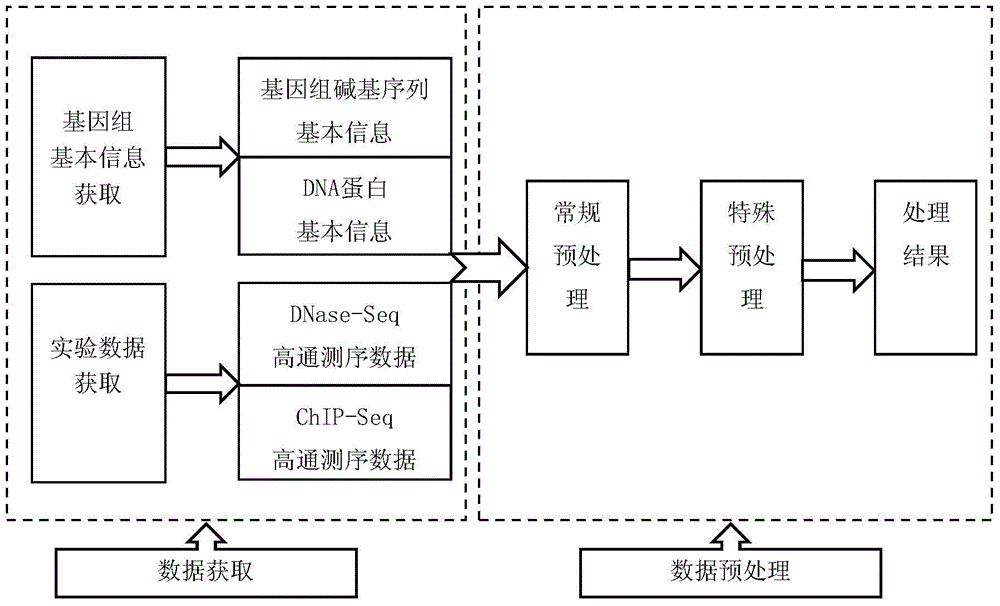
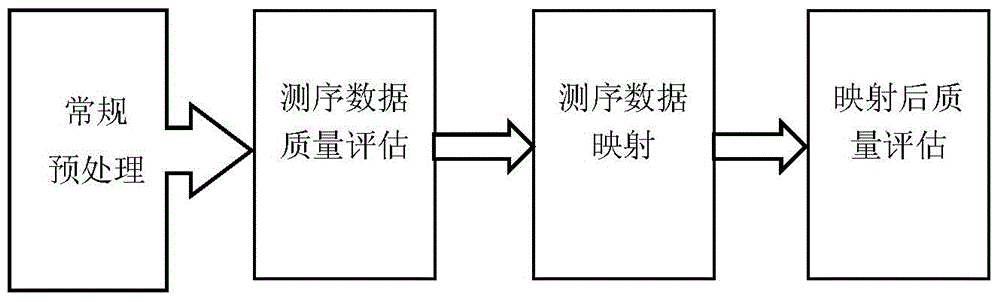
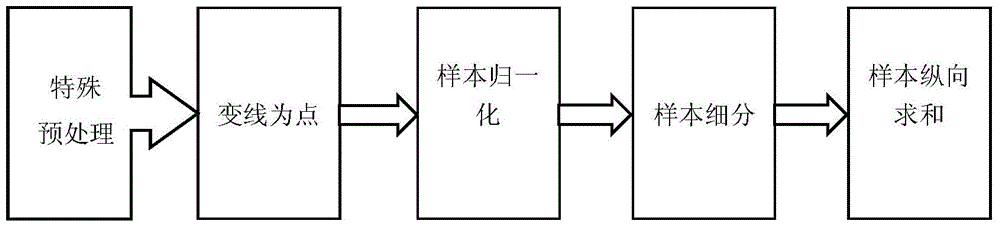
[0031] Obtain the DNase high-throughput sequencing detection data of DNA protein binding sites, conduct in-depth analysis and targeted processing, and finally achieve the purpose of highlighting the detection information of protein binding sites. like figure 1 As shown, it specifically includes the following steps:
[0032] 1. Data Acquisition
[0033]We obtain the basic information of genome base sequence from the international biological information website UCSC; obtain the basic information of DNA protein from the TRANSFAC database of BIOBASE company. The DNase-Seq and ChIP-Seq detection data of DNA protein binding sites are all from the data generated by the ENCODE project published on the UCSC website. Among them, the DNase-Seq detection data was downloaded from the data provided by the DUKE laboratory and the UW laboratory under hg19 conditions on th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com