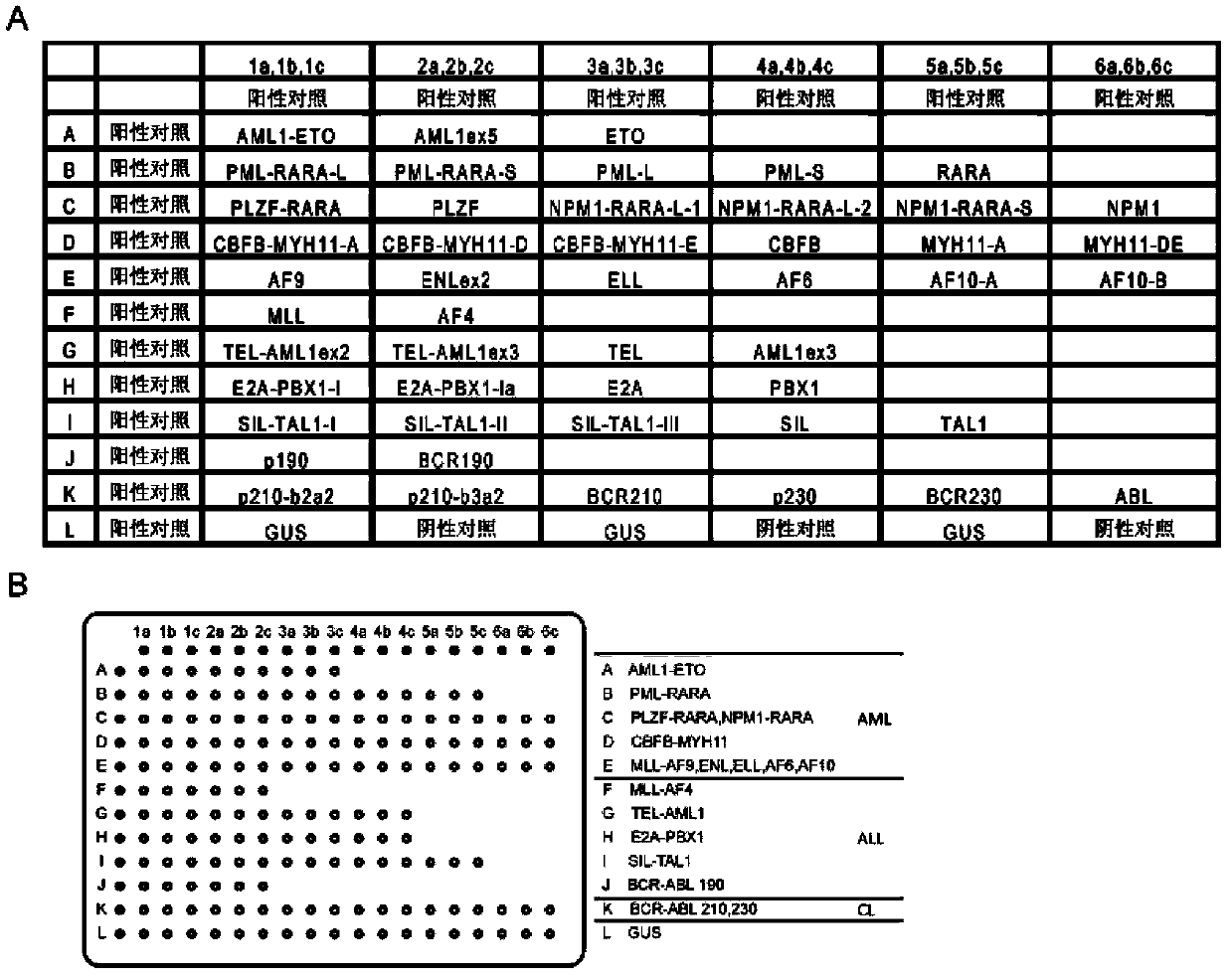
Detection method for leukemia fusion genes, primers, probes and gene chip thereof
A fusion gene and gene chip technology, applied in the field of specific probes, multiple RT-PCR combined with gene chips to detect leukemia fusion genes, primers and gene chips, can solve problems such as unfavorable clinical screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Detection of leukemia fusion gene in K-562 cell line by multiplex RT-PCR combined with gene chip
[0025] 1. Total Cell RNA Extraction
[0026] Take the K-562 cell line, use the method of Trizol, and extract the total RNA of the cells according to the instructions of Trizol of Invitrogen Company. The specific steps of extraction are as follows:
[0027] (1) Take 400 μl of the specimen stored in Trizol, add 400 μl of chloroform, mix well, and let stand at room temperature for 5 minutes;
[0028] (2) Centrifuge at 15,000 rpm for 10 minutes, and absorb the colorless supernatant after centrifugation;
[0029] (3) Add an equal volume of pre-cooled isopropanol to the above supernatant, mix it upside down and place it at room temperature for 10 minutes;
[0030] (4) Centrifuge at 15,000 rpm for 10 minutes, and discard the supernatant;
[0031] (5) Add 300 μl pre-cooled 70% ethanol to the above centrifuge tube, centrifuge at 15,000 rpm for 5 minutes, discard the s...
Embodiment 2
[0060] Embodiment 2 multiplex RT-PCR detection result
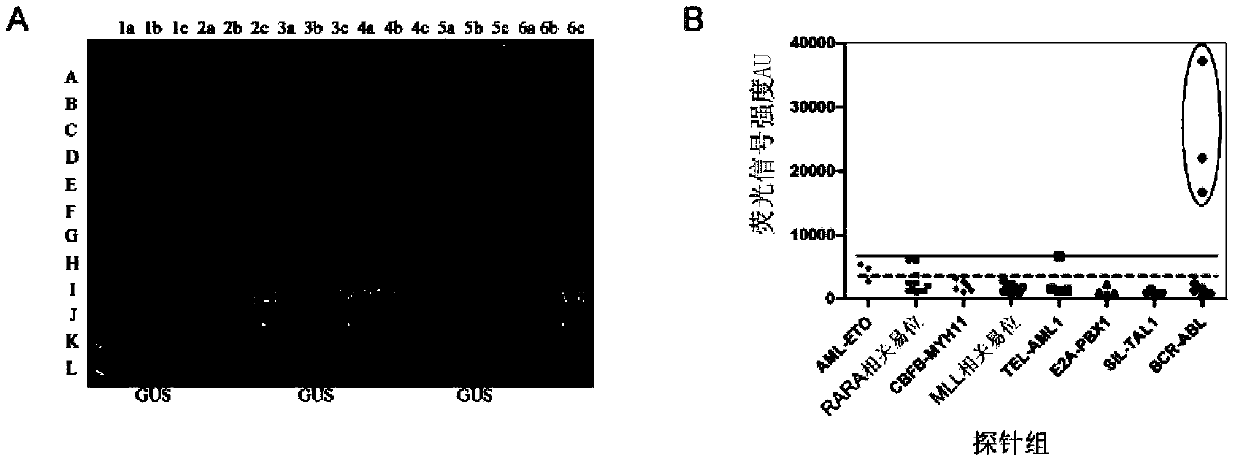
[0061] Take KASUMI-1, NB4, ME-1, THP-1, REH and K-562 leukemia cell lines, and t(15;17)PML-RARA S type, t(4;11)MLL-AF4, t(11 ;19)MLL-ENL, t(11;19)MLL-ELL, t(6;11)MLL-AF6, t(10;11)MLL-AF10, t(1;19)E2A-PBX1, t(9 22) For BCR-ABL p190 positive samples, perform the extraction of total cellular RNA, specific reverse transcription reaction and multiplex PCR amplification test according to Step 1 to Step 3 in Example 1. After the end of the test, get 3 μl of PCR product and detect it with 2% agarose gel, the results of electrophoresis are shown in image 3 .
[0062] image 3 KASUMI-1, NB4, ME-1, THP-1, REH, and K-562 leukemia cell lines are shown, along with t(15;17)PML-RARA type S, t(4;11)MLL-AF4, t (11;19)MLL-ENL, t(11;19)MLL-ELL, t(6;11)MLL-AF6, t(10;11)MLL-AF10, t(1;19)E2A-PBX1, t (9;22) Multiplex RT-PCR results of BCR-ABL p190 positive samples. The band at 328bp is the internal reference gene GUS gene, and the other b...
Embodiment 3
[0063] Embodiment 3 Sensitivity detection
[0064] Taking KASUMI-1 cells as an example, the sensitivity experiment was carried out. Starting from 2 μg of initial RNA, carry out 10-fold serial dilution with RNA of HL-60 (negative leukemia cells), and the lowest dilution is 10 -4 . Multiplex RT-PCR combined with gene chip detection to determine the sensitivity of KASUMI-1 cells, see the results Figure 4 .
[0065] Figure 4 Gel electrophoresis and chip hybridization of different dilution gradients of KASUMI-1 are shown in . It can be seen from the results of gel electrophoresis and chip hybridization that the detection sensitivity of KASUMI-1 is within 10 -3 , that is, as long as there is one cell with the AML1-ETO fusion gene in 1000 cells, it can be obtained by the method of the present invention.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com