Novel promoter and use thereof
A promoter and nucleotide sequence technology, applied in the direction of activity regulation, introduction of foreign genetic material using vectors, biofuels, etc., can solve the problems of insufficient expression level of downstream genes, achieve excellent productivity, increase productivity, and improve expression level Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
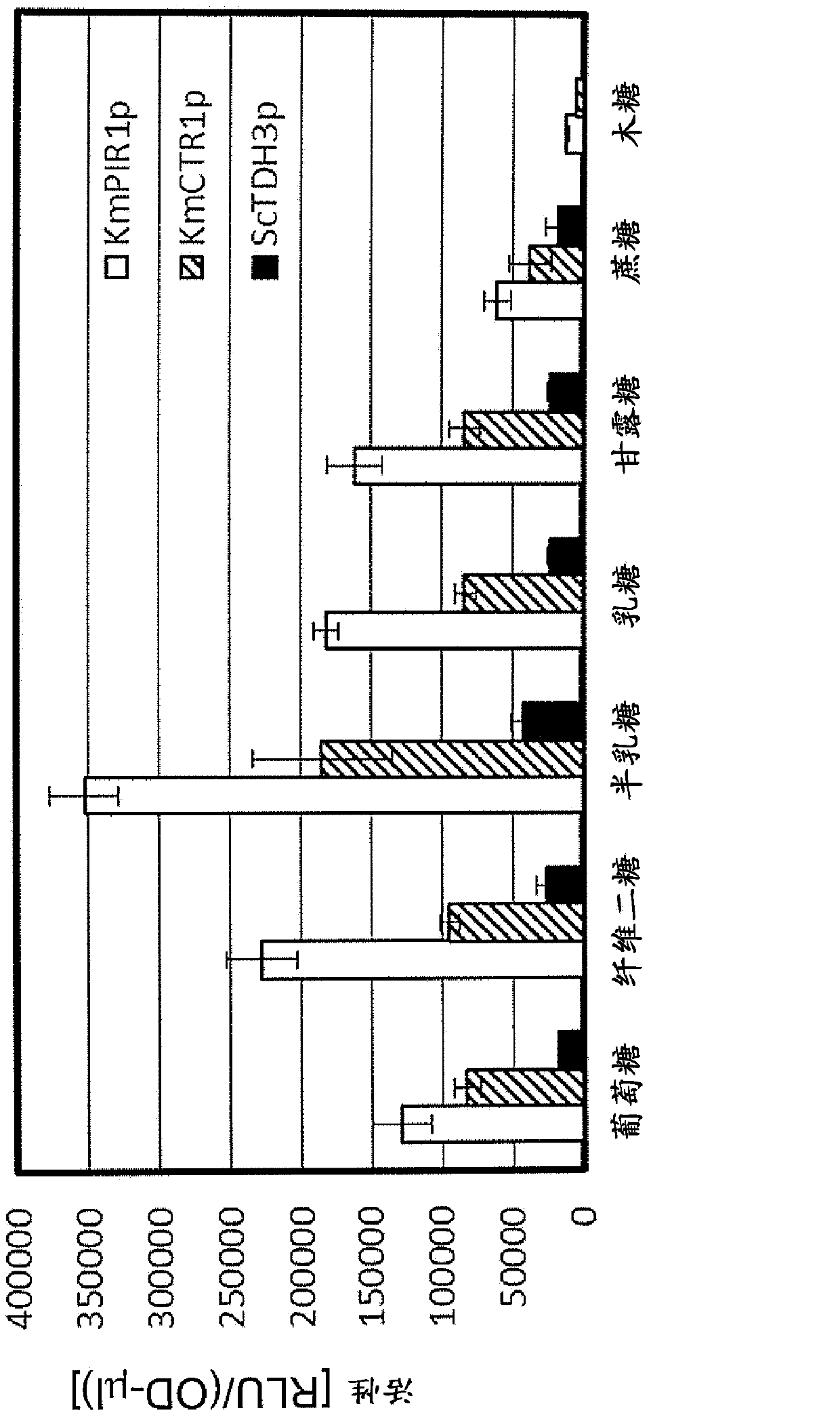
[0079] In this example, the PIR1 gene promoter and the CTR1 gene promoter in Kluyveromyces marxii, which is a thermotolerant yeast, were isolated and evaluated for promoter activity by a reporter assay method.
[0080] First, chromosomal DNA of K. marxii was prepared according to standard methods. Using the obtained chromosomal DNA as a template, KmCTR1-1085 (TAGGATCAGGAGACAATCGATATTA (SEQ ID NO: 3)) and CLuc+30c-KmCTR1-1c2
[0081] (caaagcgacagccaagatcaaggtcttcatCTTGATTGTTCAATTGTCAATTGTC (SEQ ID NO: 4)) as a primer, and KOD plus DNA polymerase (Toyobo), PCR was performed. After PCR is completed, the reaction solution is subjected to electrophoresis to obtain the target DNA band.
[0082] In addition, chromosomal DNA of Saccharomyces cerevisiae RAK4296 strain (MATa his3 200leu2 0met15 0trp1-delta-63ura3 0::ScGAL10p-yCLuc-15C-LEU2) was prepared according to a standard method. Using the obtained chromosomal DNA as a template, yCLuc+1 (ATGAAGACCTTGATCTTGGC (SEQ ID NO: 5)) and U...
Embodiment 2
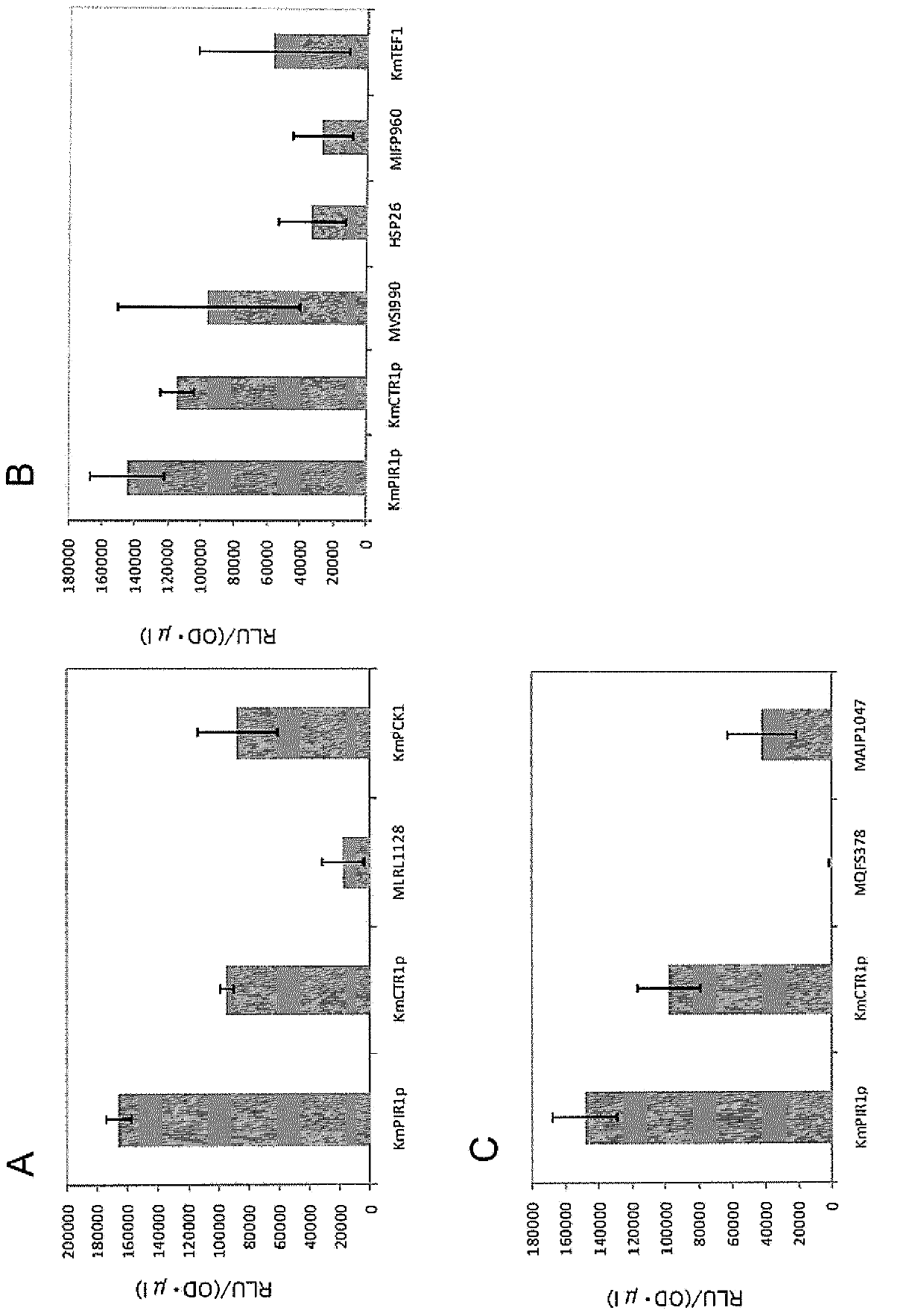
[0099] In this example, the PIR1 promoter of Kluyveromyces marxii and the CTR1 promoter of Kluyveromyces markeks evaluated in Example 1 were examined in terms of functional regions.
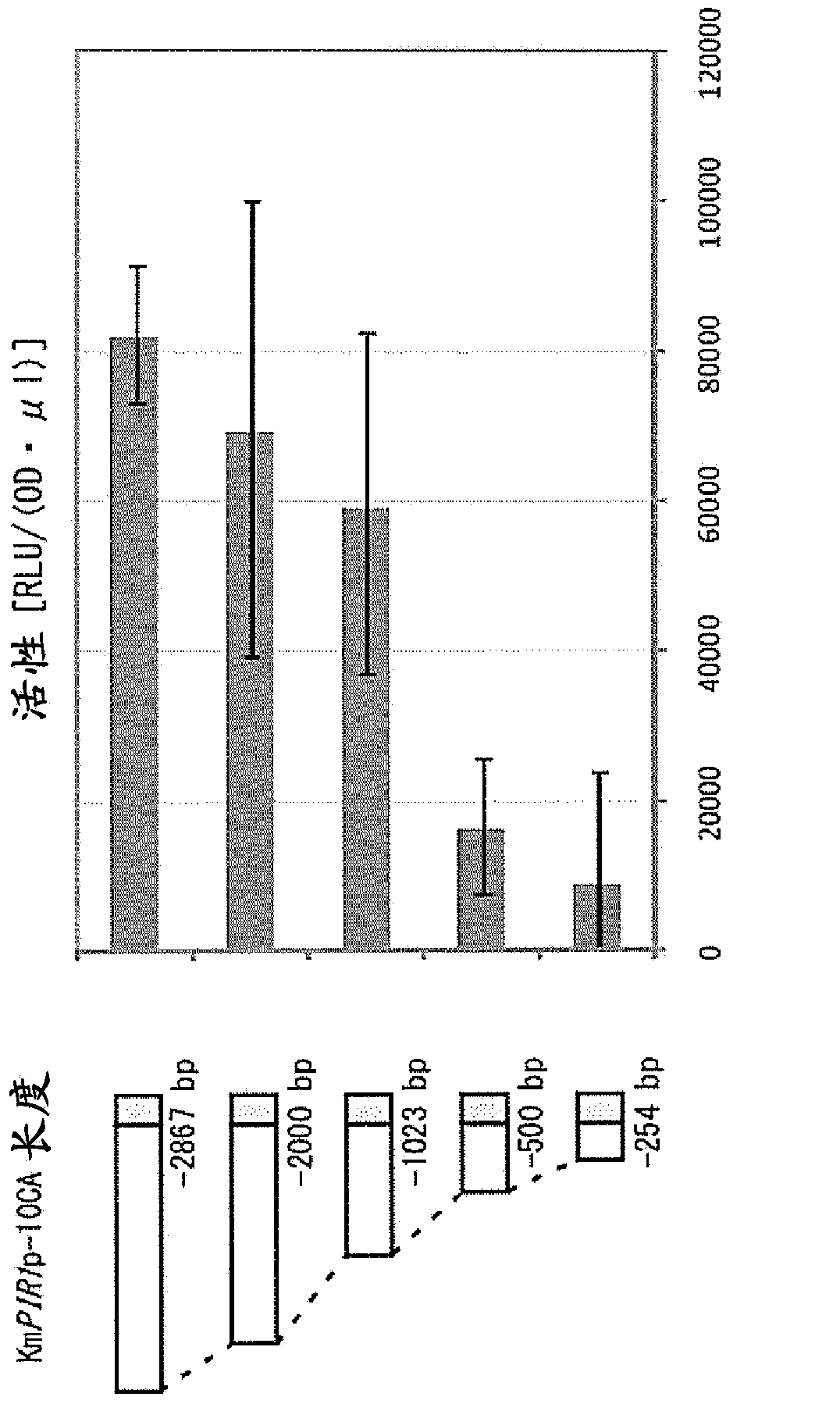
[0100] Specifically, PCR was performed using the chromosomal DNA of the RAK5689 strain prepared in Example 1 as a template, the URA3-280c primer, and each of the primers listed in Table 2. That is, as a result of PCR, DNA fragments each containing a luciferase gene having a different characteristic from that obtained by cutting the PIR1 promoter evaluated in Example 1 on the 5' terminal side were synthesized. One of the DNA fragments of different lengths is fused. Each PCR-amplified fragment was introduced into K. marxense strain RAK4174, and luciferase activity was measured as described in Example 1.
[0101] [Table 2]
[0102] KmPIR1-2867
[0103] image 3 Displays the length of the DNA fragment evaluated and the luciferase activity measurement. Such as image 3 As shown in , i...
Embodiment 3
[0109] In this example, the PIR1 promoter and the CTR1 promoter of Kluyveromyces marxii evaluated in Example 1 were used as promoters for introducing foreign genes into thermotolerant yeast to evaluate their activation child activity.
[0110][Generation of xyl1 / xyl2 double deletion mutant transfected with XI gene]
[0111] The xyl2 gene used to delete the RAK6163xyl1 deletion strain (xyl1::ScURA3,leu2,his3) from the Kluyveromyces marxense strain DMKU3-1042 (also called "RRAK35966") was prepared as described below and simultaneously introduced xylose isomer Carrier of the enzyme (XI) gene. The XI gene used herein is the RsXI-C1-opt (hereinafter referred to as "RsXI") gene prepared by fully synthesizing the following gene while adjusting the codon usage frequency relative to the codon usage frequency in yeast: Japanese Patent Publication (Kokai ) No. 2011-147445A, the XI gene from the intestinal symbiotic protist of Reticulitermes speratus is known to show activity in Sacchar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com