Beta-galactosidase combined mutant with high transglycosylation activity as well as preparation method and application of beta-galactosidase combined mutant
A technology of galactosidase and high transglycosidase is applied in the field of β-galactosidase combination mutants and their preparation, and can solve the problems of low yield, long reaction time, low galacto-oligosaccharide synthesis yield and the like, and achieves the High-efficiency transglycosidic activity, effect of high transglycosidic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
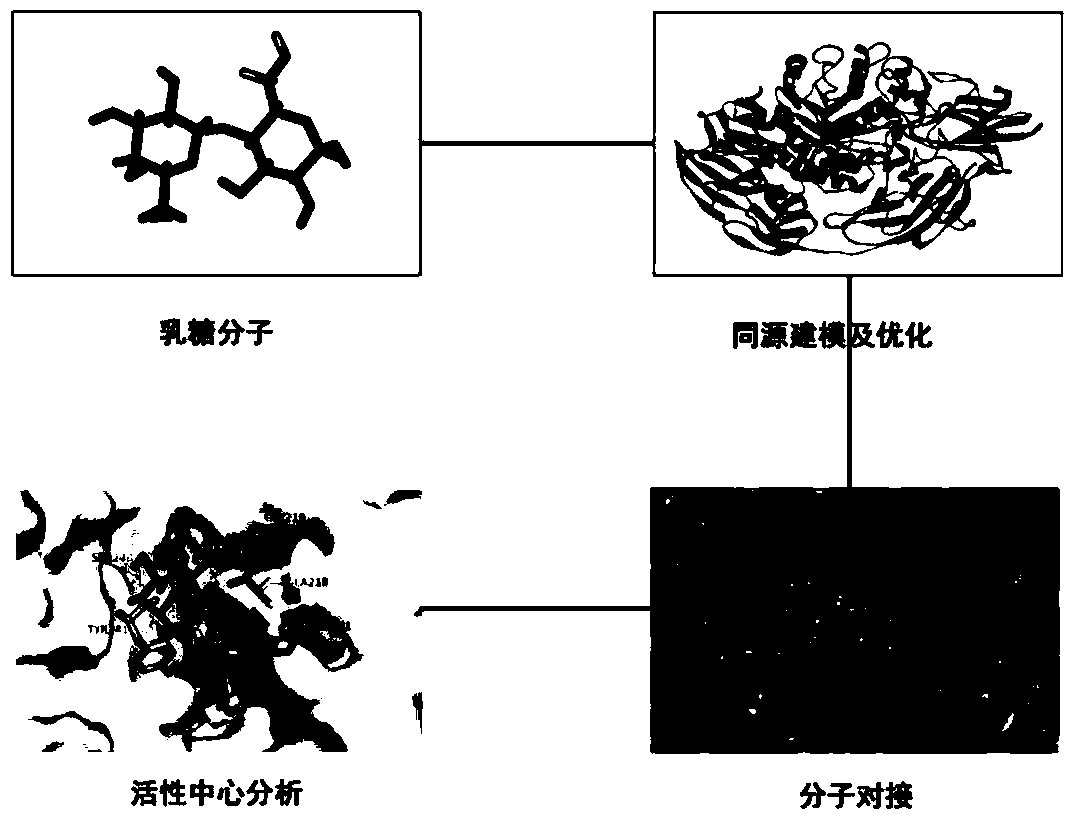
[0034] Example 1: Prediction of tertiary structure and mutation site of β-galactosidase
[0035] The β-galactosidase gene lacb' that removes its own signal peptide is cloned from Aspergillus candidus in our laboratory. The gene that removes its own signal peptide sequence consists of 2958 nucleotides. The specific sequence is as follows: As shown in 1, the protein encoded by the gene consists of 986 amino acids, and the specific sequence is shown in sequence 2.
[0036] The β-galactosidase gene laco', which removes its own signal peptide, was cloned from Aspergillus oryze in our laboratory, and it also consists of 2958 nucleotides. The specific sequence is shown in sequence 3. The gene The encoded protein also consists of 986 amino acids, and the specific sequence is shown in SEQ ID NO:4. There are only three amino acid differences between its amino acid sequence and the protein encoded by the lacbˊ gene: at position 231: lacbˊ(Gly), lacoˊ(Ser); at position 401: lacbˊ(Met), l...
Embodiment 2
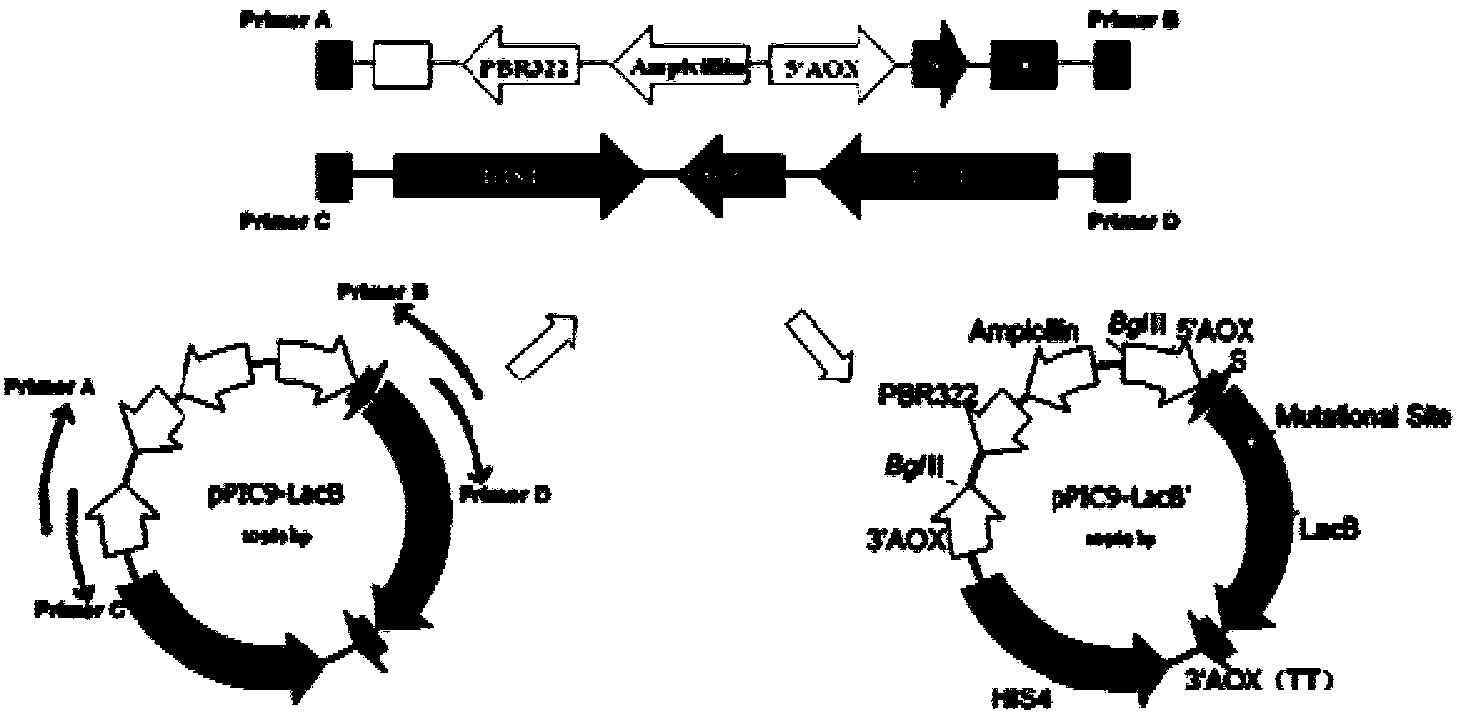
[0041] Example 2: Construction of Pichia pastoris single point saturation mutant library
[0042] 1. Materials and methods
[0043] (1) Strain and carrier
[0044] The wild-type gene is derived from the β-galactosidase gene lacb' of Aspergillus leucobacter that removed its own signal peptide, and the β-galactosidase gene laco' from Aspergillus oryzae, which were cloned in the previous stage of our laboratory. The specific sequence is as follows: 1 and Sequence 3, linked to pPIC9 expression vector, and expressed in Pichia pastoris GS115; Escherichia coli Trans1-T1 competent cells were purchased from TransGen; pPIC9 expression vector, Pichia GS115 was purchased from Invitrogen.
[0045] (2) Preparation of medium and related solutions
[0046] For Pichia pastoris transformation, culture and screening conventional media and reagents, refer to the instructions of Invitrogen.
[0047] PTM trace salt: 0.6% CuSO 4 , 0.008% NaI 2 , 0.3% MnSO 4 , 0.02% Na 2 MoO 4 , 0.002%H 3 BO...
Embodiment 3
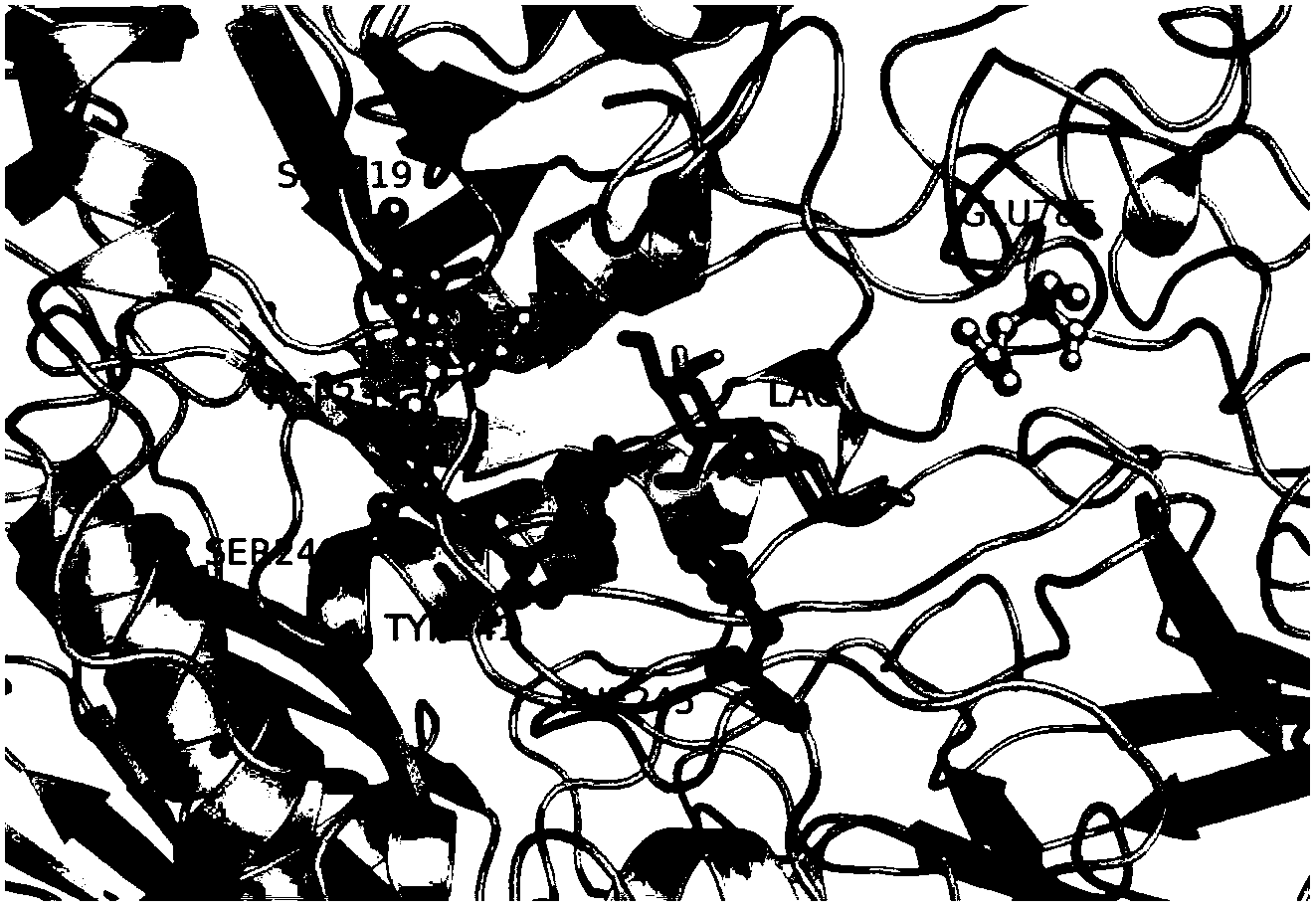
[0077] Example 3: Screening of S219 Saturation Mutant Library and Synthesis of Oligosaccharides
[0078] 200 positive Pichia pastoris clones from the S219 mutant library were selected to measure their transglycoside activity (oligosaccharide production), and their nucleotide sequences were determined. Sequencing showed that these mutants were mutated to 8 different amino acids, all of which could improve the transglycosidic activity of the mutant enzyme (see Table 3), especially amino acids with smaller side chains such as Gly, Ala, Val and negatively charged The polar amino acid Glu is more prominent, and the mutation to Gly is the most obvious (see attached Figure 4 ), the production of oligosaccharides can be increased by 26.6%. After mutation to small side chain and negatively charged Glu, the production of oligosaccharides increased by 25.7%. When S219 was mutated to Ala and Val, the production of oligosaccharides increased by 15.0% and 15.5%, respectively. Mutated to A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com