Connected sequencing method for DNA by coupling and coding two rounds of signals
A DNA sequencing and signal coupling technology, applied in the biological field, can solve problems such as low efficiency, reduced sequencing accuracy, and increased sequencing equipment costs, achieving the effects of reducing precision requirements, increasing sequencing speed, and reducing sequencing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] A DNA ligation and sequencing method for two-round signal coupling coding, specifically comprising the following steps:
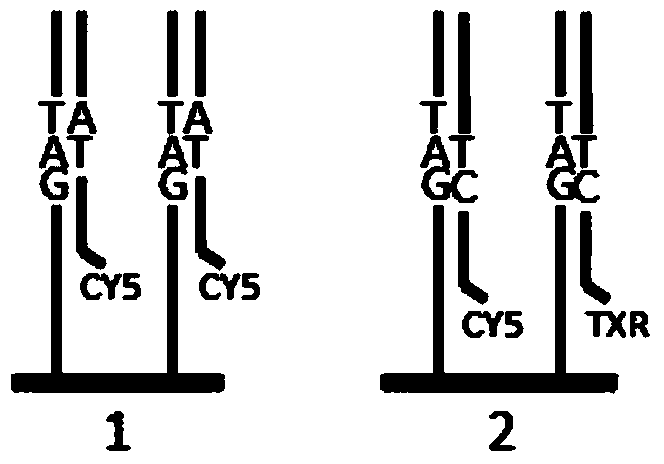
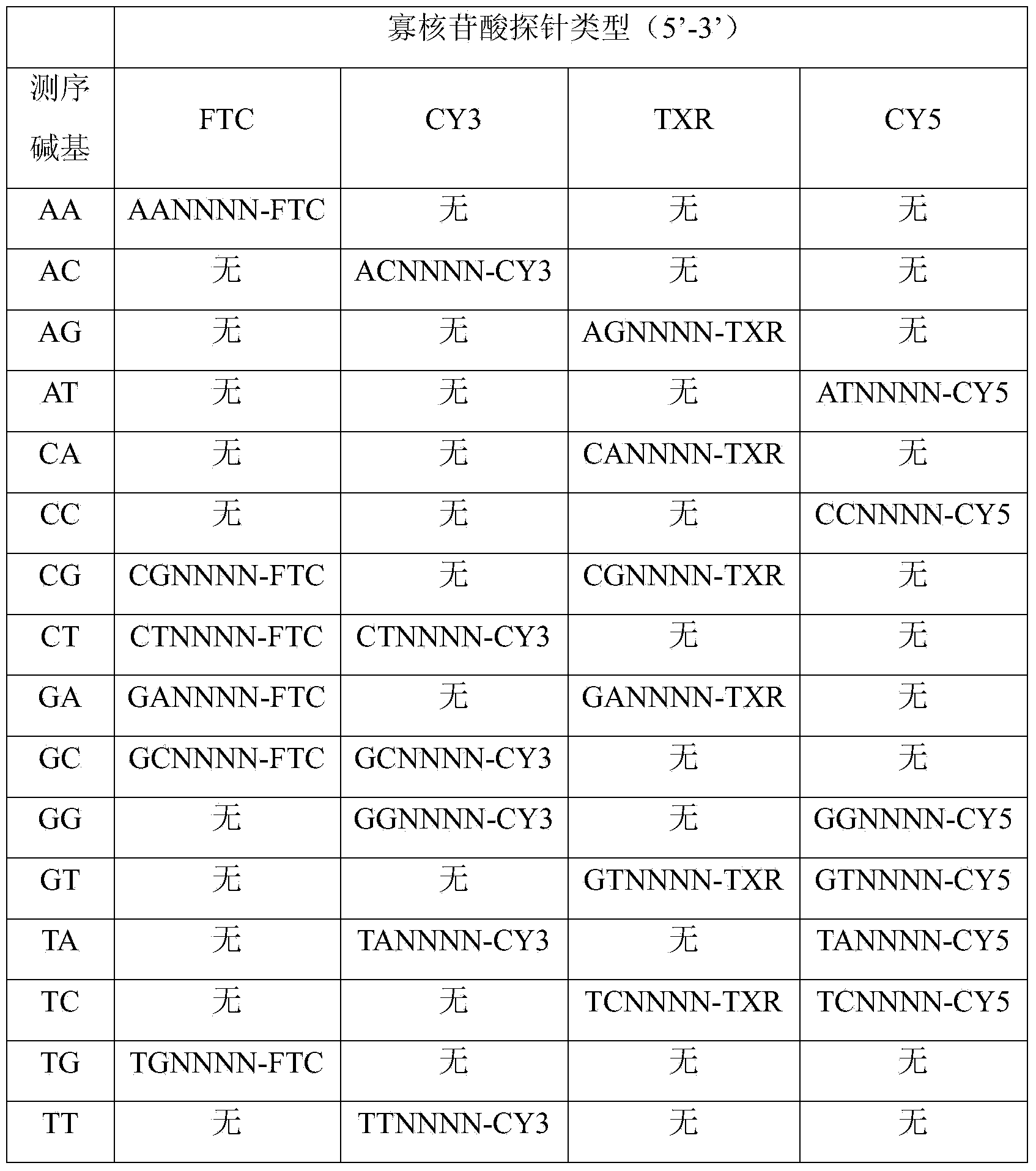
[0038] Step 1, prepare a set of coupled-encoded DNA sequencing probes: there are 16 DNA sequencing probes in total, corresponding to 16 combinations of two-digit sequencing bases; one marker or any two of the four markers are used. The markers are combined to form a total of 10 labeling states, and 8 of the 10 labeling states are selected to label 16 sequencing probes, and each labeling state corresponds to two sequencing probes;
[0039] Step 2, two rounds of sequencing signals are coupled to determine the three-digit base sequence information: using a set of sequencing probes obtained in step 1, the first round of sequencing The two bases to be tested are complementary paired with one of the two sequenced bases corresponding to the state of the marker; a second round of sequencing reactions is performed, and the second round of sequencing reactions...
Embodiment 2
[0049] Two rounds of signal-coupling coding sequencing method (respectively labeling scheme) detected a total of 32 base pairs of sequences from 45332-45363 on the RP11-354P11 clone on human chromosome 17;
[0050] Human whole genome DNA was extracted, and the target fragment was amplified by PCR method. The primers were P1 and P2 (see Table 3 for the sequence), and the 5' end of the primer P1 was modified with a hydroxyl group. After the amplified PCR product was immobilized on the surface of the aldehyde group-modified glass slide, the glass slide was heated to 95°C to detach one strand of the double-stranded PCR product from the immobilized strand, and then the sequencing reaction was started.
[0051] Relevant oligonucleotide sequence information in Table 3 Example 1
[0052] sequence name
sequence information
sequence to be tested
5’-CACGGACCAGCTGCCCTGGACCAGCTGCAAGA-3’
P1
OH-5’-CGCTATACTACCTCATCTCCTCCTTCACG-3’
P2
5'-GCAGTTGCCA...
Embodiment 3
[0062] Two rounds of signal-coupled coding sequencing method (common labeling scheme) detected a total of 32 base pairs of sequences from 45332-45363 on the RP11-354P11 clone on human chromosome 17;
[0063] The human whole genome DNA was extracted, and the target fragment was amplified by PCR method. The primers were P1 and P2 (see Table 5 for the sequence), and the 5' end of the primer P1 was modified with a hydroxyl group. After the amplified PCR product was immobilized on the surface of the aldehyde group-modified glass slide, the glass slide was heated to 95°C to detach one strand of the double-stranded PCR product from the immobilized strand, and then the sequencing reaction was started.
[0064] Related oligonucleotide sequence information in table 5 example 2
[0065] sequence name
sequence information
sequence to be tested
5’-CACGGACCAGCTGCCCTGGACCAGCTGCAAGA-3’
P1
OH-5’-CGCTATACTACCTCATCTCCTCCTTCACG-3’
P2
5'-GCAGTTGCCAGTGT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com