Intron-source circular RNA molecules and application of cyclization key nucleotide sequences of intron-source circular RNA molecules
A nucleic acid sequence and intron technology, applied in the field of nucleic acid research, can solve the problems of no reports, and the discovery of key nucleic acid sequences that have no reports on the looping function, etc., to achieve the effect of enriching knowledge
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
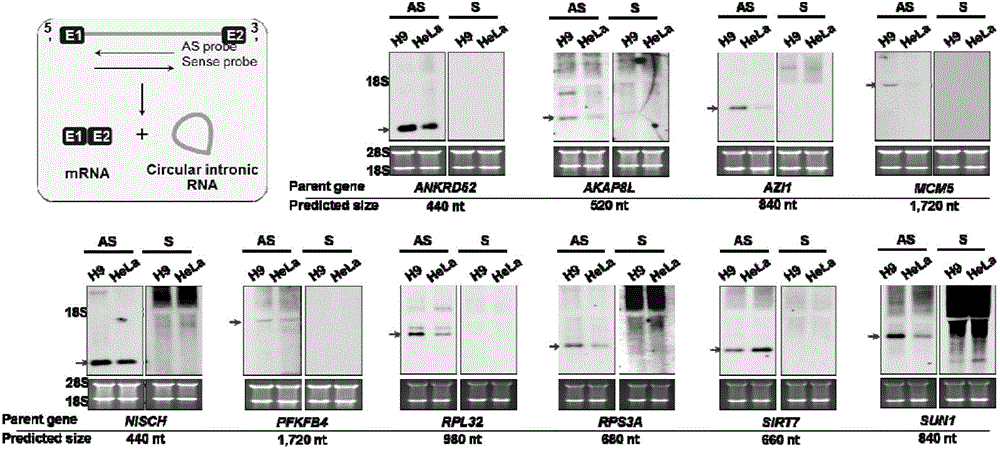
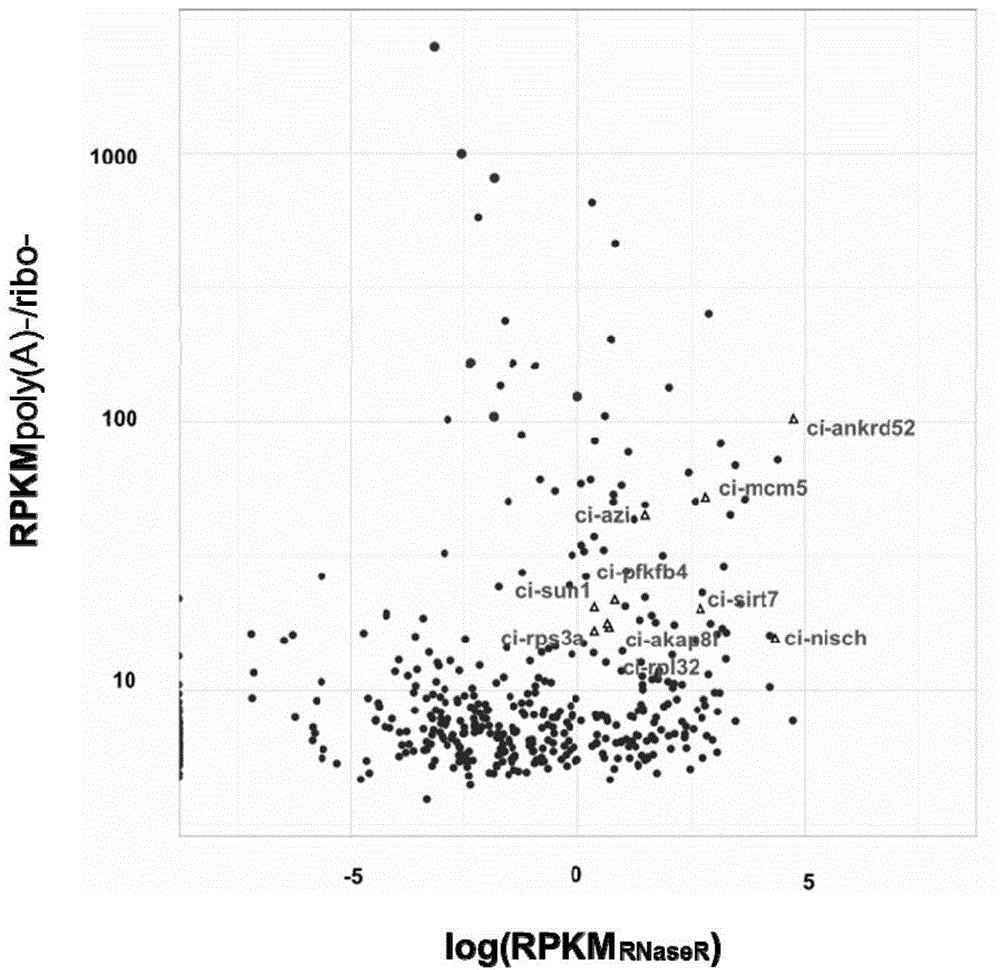
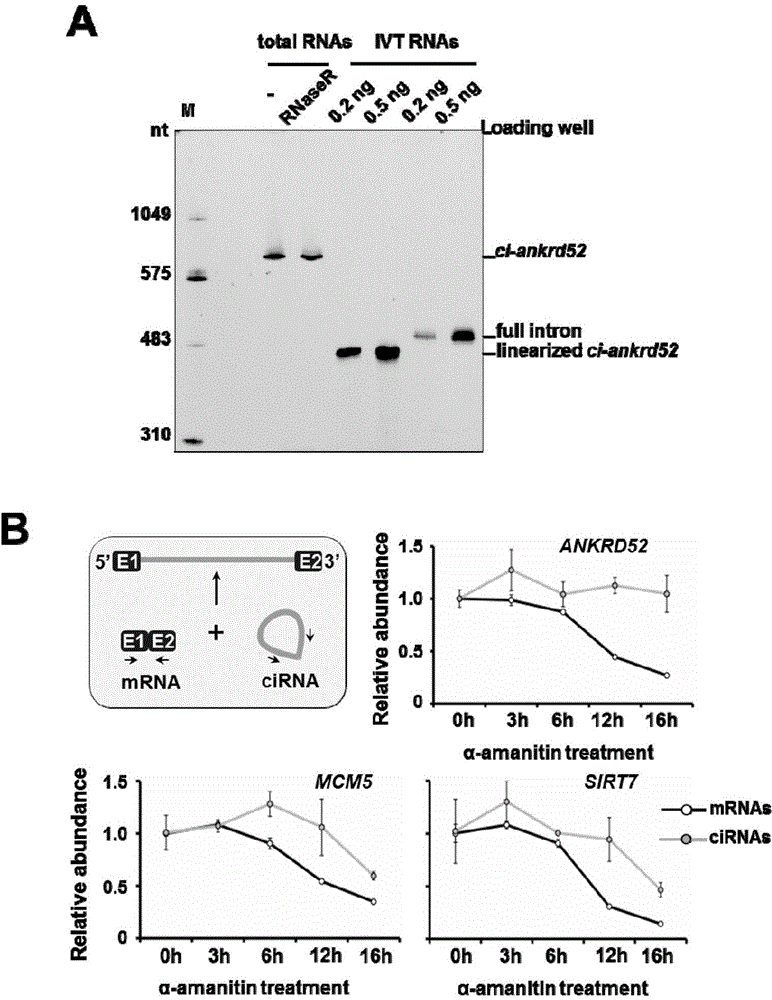
[0036] Example 1 Discovery and Identification of Intron-derived Circular RNA
[0037] 1. Experimental materials
[0038] 1) Reagents
[0039] TRIzol, oligo(dT) magnetic beads, RiboMinus kit, and CDP-STAR reagent were purchased from Invitrogen; Dig wash and block buffer set, 10×Dig RNA labeling mix, Anti-Digoxigenin, Fab Fragment were purchased from Roche; AmpliScribe TM T7, T3, and SP6 High Yield Transcription Kits, RNase R were purchased from Epicentre; Illumina TruSeq Stranded Total RNA HT Sample Prep Kit was purchased from Illumina; RNA Reverse Kit was purchased from Takara; 2×QPCR mix was purchased from Toyobo; DNase Ⅰ was purchased from Ambion Company; EcoR Ⅰ and Not Ⅰ were purchased from NEB Company; α-amanitin was purchased from Sigma Company.
[0040] 2) Cell lines and vectors
[0041] Stem cells H9 were cultured on Martigel-coated culture dishes using CM (fibroblast-conditioned medium) medium, which was MEF supplemented with 4 ng / mL fibroblast growth factor (bFGF,...
Embodiment 2
[0087] Example 2 Discovery and Identification of Key Nucleic Acid Sequences for Circular RNA from Intron Origin
[0088] 1. Reagents
[0089] RNA Marker Ⅲ was purchased from Roche Company; Nhe Ⅰ, BamH Ⅰ, Hind Ⅲ were purchased from NEB Company; agarose gel recovery kit, common DNA product recovery kit were purchased from Tiangen Company.
[0090] 2. Cell lines and vectors
[0091] PA-1 cells were adherently cultured in MEMα medium (Gibco) supplemented with 10% fetal bovine serum (Gbico), 1% L-glutamine, and 1‰ double antibody. The vector used to construct the plasmid is pcDNA3 and the modified pEGFP-C1 plasmid, namely pZW1, which divides egfp into two parts, with a splicing site and a restriction site in the middle. Green fluorescence formation (Systematic Identification and Analysis of Exonic Splicing Silencers, Wang et al., 2004).
[0092] 3. Experimental method
[0093] 3.1 Reconstruction of ciRNAs
[0094] 3.1.1 Construction of expression plasmid
[0095] In order to ...
Embodiment 3
[0126] Example 3 New materials and new methods for looping key sequences to induce looping of linear RNA molecules
[0127] 1. Reagents
[0128] Nhe Ⅰ, BamH Ⅰ, Hind Ⅲ were purchased from NEB Company; QuikChange Site-Directed Mutagenesis Kit was purchased from Stratagene Company; Agarose Gel Recovery Kit and Common DNA Product Recovery Kit were purchased from Tiangen Company; AmpliScribe TM T7, T3, and SP6 High Yield Transcription Kits were purchased from Epicentre.
[0129] 2. Cell lines and vectors
[0130] PA-1 cells were adherently cultured in MEMα medium (Gibco) supplemented with 10% fetal bovine serum (Gbico), 1% L-glutamine, and 1‰ double antibody. The vector is pcDNA3 and pZW1 after transformation.
[0131] 3. Experimental method
[0132] The purpose of this embodiment is to explore whether the key sequence of ciRNAs has wide applicability. The inventor selected an intron that cannot form circular RNA under natural conditions, that is, the fifth intron (intron5) of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com