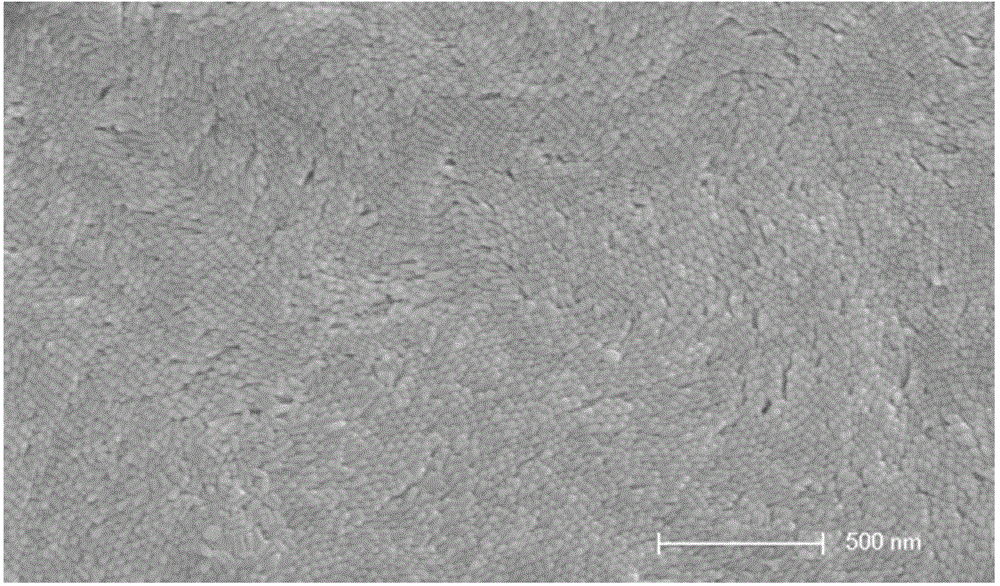
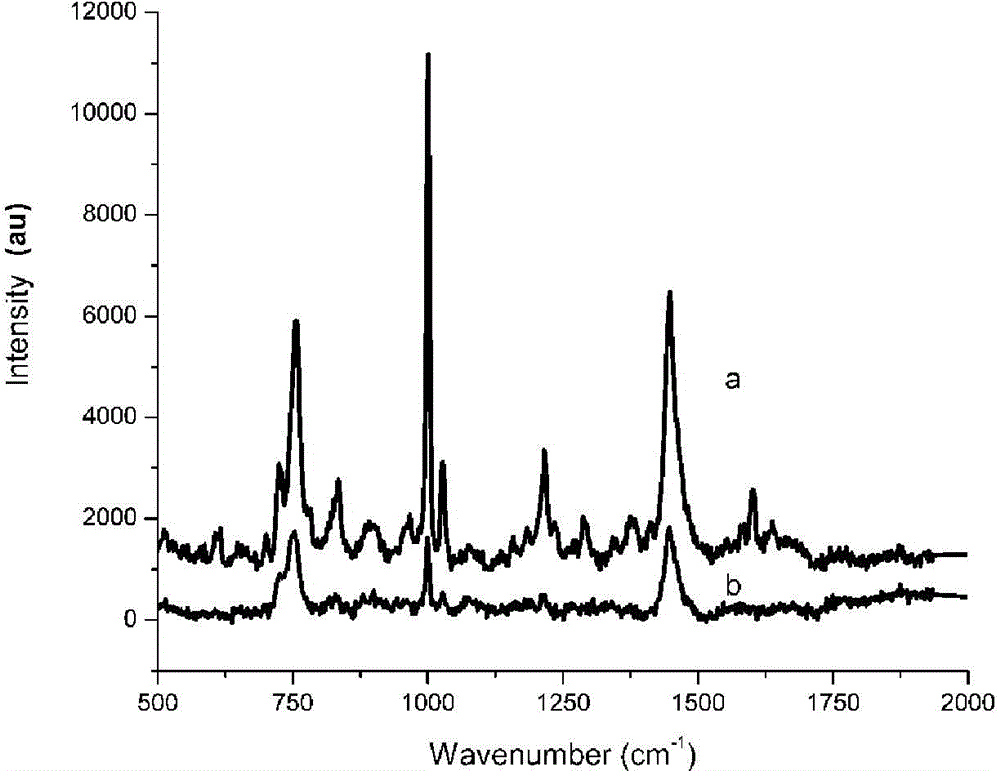
Method for enhancing Raman signals with nano super-crystal technology to identify microorganisms
A Raman signal and microorganism technology, applied in Raman scattering, material excitation analysis, etc., can solve the problems of long detection period, inaccurate detection, unable to meet the requirements of rapid and accurate identification of microorganisms, and achieve rapid identification and simple identification methods. , the effect of broad universality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Recognition of Escherichia coli by enhancing the intrinsic Raman signal of Escherichia coli through three-dimensional supercrystals of gold-silver core-shell nanorods
[0027] In this embodiment, the intrinsic Raman signal of Escherichia coli is enhanced by three-dimensional supercrystals of gold-silver core-shell nanorods to identify Escherichia coli, and the procedure is as follows:
[0028] Step 1: Dissolve 20 mL of 0.5 mM auric acid chloride (HAuCl 4 ) solution and 20mL, 0.2M cetyltrimethylammonium bromide (CTAB) were mixed under ice bath conditions to obtain a mixed solution; 1.2mL, 0.02M sodium borohydride (NaBH 4 ) into the mixed solution, under the condition of 1000rpm rotating speed, vigorously stir for 2 minutes, and stand at 25°C for 1 hour to obtain a solution containing gold nano-seeds;
[0029] Mix 200mL, 0.2M CTAB with 10mL, 4mM silver nitrate (AgNO 3 ) solutions were mixed at room temperature to form a yellow growth solution; 200mL, 1mM HAuC...
Embodiment 2
[0039] Example 2: Enhancing Raman signal to identify yeast by three-dimensional supercrystal of golden triangle nanosheet
[0040] In this embodiment, the Raman signal is enhanced by the three-dimensional supercrystal of the golden triangle nanosheet to identify the specific steps of the yeast:
[0041] Step 1: Dilute 1mL, 0.01M chloroauric acid solution and 1mL, 0.01M trisodium citrate solution with water to 40mL, mix under ice bath conditions to obtain a mixed solution, then add 1mL, 0.1M sodium borohydride, and rotate at 1000rpm Under the conditions, stir vigorously for 2 minutes, and stand at 25° C. for 2-4 hours to obtain a solution containing gold nanoparticles.
[0042] Then, 225mL, 0.05M CTAB, 1mL, 0.1M KI, 1.25mL, 0.1M ascorbic acid, 1.25mL, 0.1M NaOH, 6.25mL, 0.01M chloroauric acid solution were mixed to form a mixed growth solution. Then, take 22.5mL from Erlenmeyer flask C containing 225mL mixed growth solution into Erlenmeyer flask B, and take 2.25mL from Erlenme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com