Kit for isothermal DNA amplification starting from an RNA template
A kit and template technology, applied in the field of synthetic deoxyribonucleic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
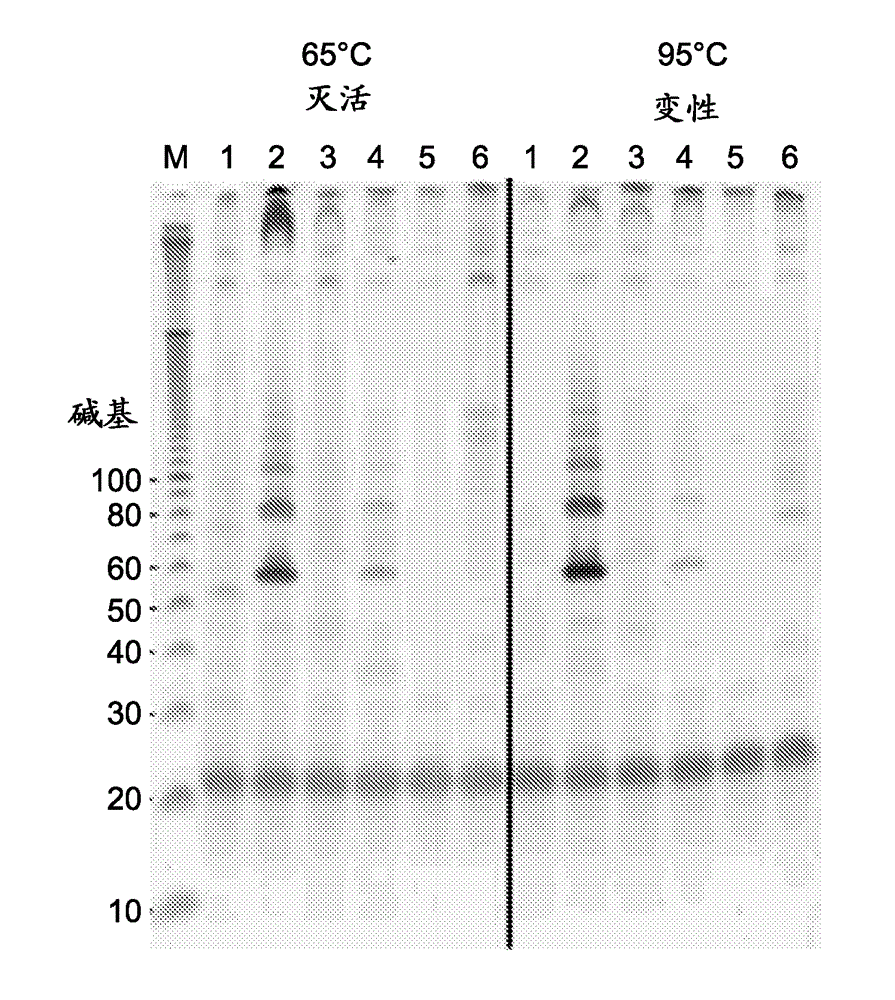
[0110] Example 1 Amplification of DNA starting from RNA template, irrespective of denaturing conditions.
[0111] First Choice™ Human Spleen Total RNA was obtained from Life Technologies™. Oligos were obtained from Integrated DNA Technologies, Inc. cDNA:RNA heteroduplexes were prepared from total spleen RNA using Oligo SEQ ID NO: 1 with SuperScript® II (SS II) reverse transcriptase and accompanying buffer purchased from Life Technologies® and prepared according to the manufacturer's instructions .
[0112] Table 1: Oligonucleotide sequences used in Example 1.
[0113] SEQ. ID No. oligonucleotide sequence 1 5'-d[CCAGCCTGGGCATCCTTGAI*T]-3' 2 5'-d[TTCAGCTCTCGGAACATCTCI*A]-3' 3 5'-d[TCACGCCCACGGATCTGAAI*G]-3' 4 5'-d[CATCCAGTGGTTTCTTCTTTI*G]-3' 5 5'-d[GAGAGGAGCTGGTGTTGTTI*G]-3' 6 5'-d[TGCTCGCTTAGTGCTCCCTI*G]-3' 7 5'-d[TTGTGCCTGTCCTGGGAGAI*A]-3' 8 5'-d[GACGGAACAGCTTTGAGGTI*C]-3' 9 5'-d[CACACTGGAAGACTCCAGTI*G]-3' ...
Embodiment 2
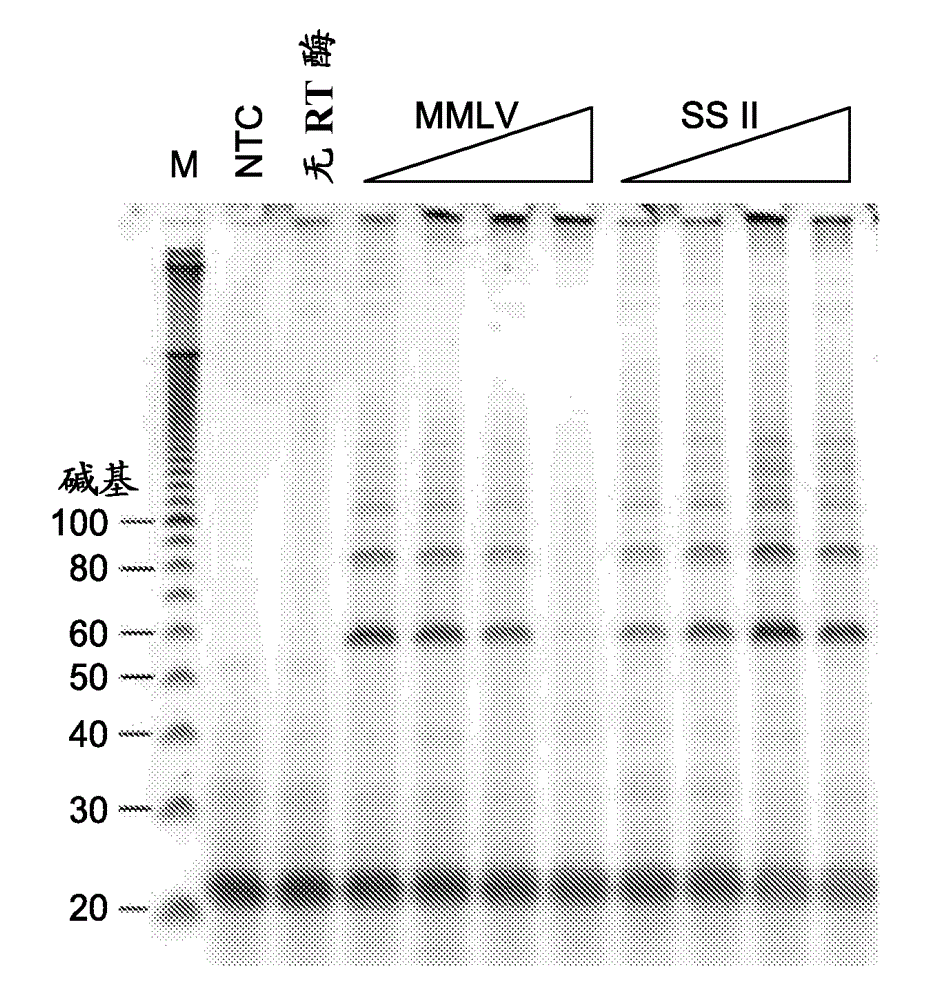
[0125] Example 2 Reverse transcription using different reverse transcriptases
[0126] Isothermal amplification reactions were prepared and analyzed as in Example 1, wherein denaturation of the starting RNA template was performed at 95°C. For this example, increasing amounts of Moloney murine leukemia virus (MMLV; Life Technologies) reverse transcriptase (RTase), which contains both reverse transcriptase and RNase H activity, or SS II RTase (reduced RNase H activity ) was added to the amplification reaction. Reactions contained 12.5, 25, 50 or 100 units of MMLV or SS II as indicated. Amplification reactions containing either RTase synthesized fragments of the expected size, indicating successful amplification of the target mRNA in a single combined reaction of RNA reverse transcription followed by amplification, as Figure 5 shown.
Embodiment 3
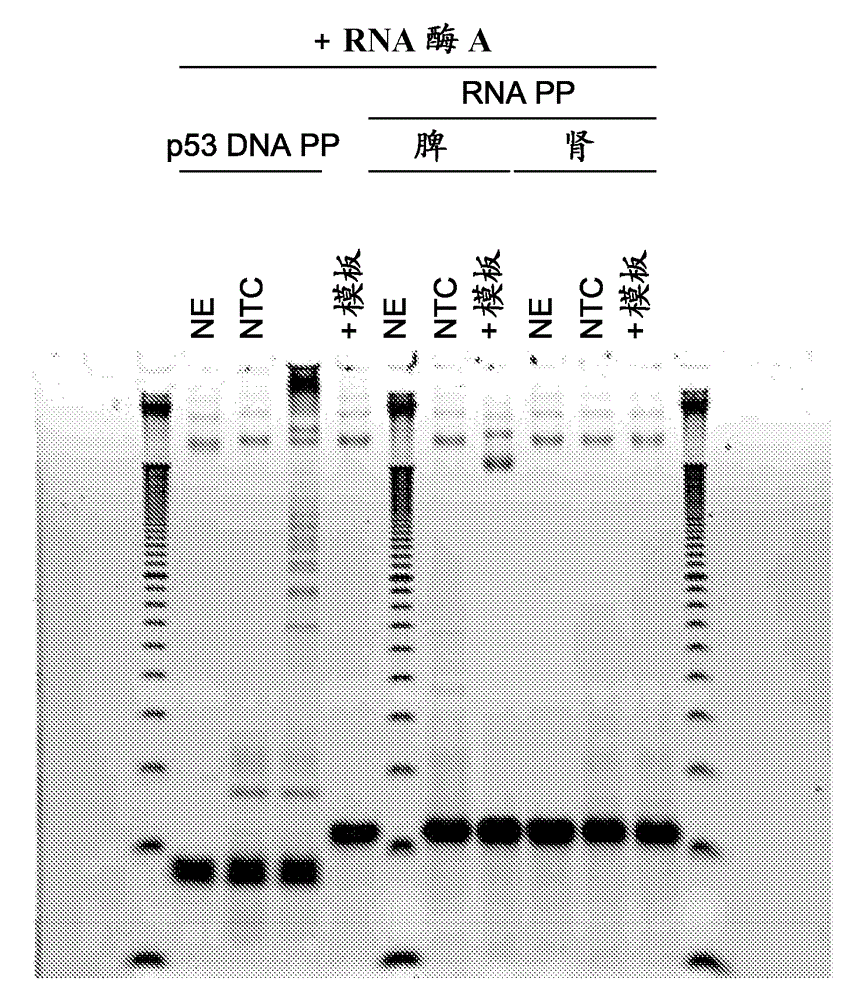
[0127] Example 3 Isothermal amplification of RNA in a single reaction mixture without denaturation
[0128] Isothermal amplification reactions were prepared and analyzed as in Example 1, but without any denaturation of the starting RNA template prior to amplification. 200 ng of total RNA (from spleen or kidney) was amplified alone with or without 30 min RNase A pretreatment (to eliminate RNA, as a negative control). The oligo mixture was the same as previously used in Example 1, which had amplified the correct product size starting from the cDNA:RNA template. RNase A was not inactivated prior to amplification. 50 units of SS II were included in all reactions.
[0129] Table 3 is used for the oligonucleotide sequence of embodiment 3
[0130]
[0131] Table 3 contains oligomers of SEQ ID No 13 - 24, wherein "I" represents inosine and "*" represents the phosphorothioate bond in the oligomer sequence. Human genomic DNA was amplified under the same conditions using differe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com