Method for developing endangered rhododendron molle SSR primer on basis of RAD-seq
A sheep slug and genome technology is applied in the field of developing endangered sheep slug SSR primers based on RAD-seq, which can solve the problems of low development efficiency, high development cost, complicated SSR labeling process, etc., and achieve the effect of fast cost and efficient method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021] The present invention is achieved like this, and described method comprises the following steps:
[0022] 1. DNA extraction and quality inspection: The modified CTAB method was used to extract the genomic DNA of the roe deer, the purity and integrity of the DNA were detected by agarose gel electrophoresis and Nanodrop, and the concentration of the genomic DNA was detected by Qubit.
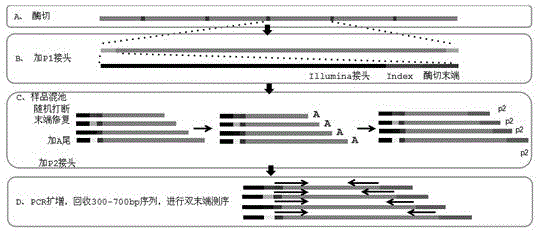
[0023] 2. Library construction: Qualified DNA samples are digested, randomly interrupted, end-repaired, A-tailed, sequenced adapters added, purified, PCR amplified, and 300-700 bp sequences recovered by agarose gel electrophoresis, etc. , which completes the genome library preparation. The schematic diagram of library construction is attached figure 1 .
[0024] 3. Library inspection: After the library is constructed, use Qubit2.0 for preliminary quantification, dilute the library to 1ng / ul, then use Agilent 2100 to detect the insert size, and then use the Q-PCR method to accurately quan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com