Target genes for detecting salmonella paratyphi, PCR primer pair as well as detection method and applications thereof
A technology for paratyphoid C and Salmonella, applied in botany equipment and methods, microbial-based methods, biochemical equipment and methods, etc., can solve problems such as long detection cycle, inaccurate detection results, and complicated detection process, and achieve The effect of shortened detection time, strong singleness, and simple result judgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
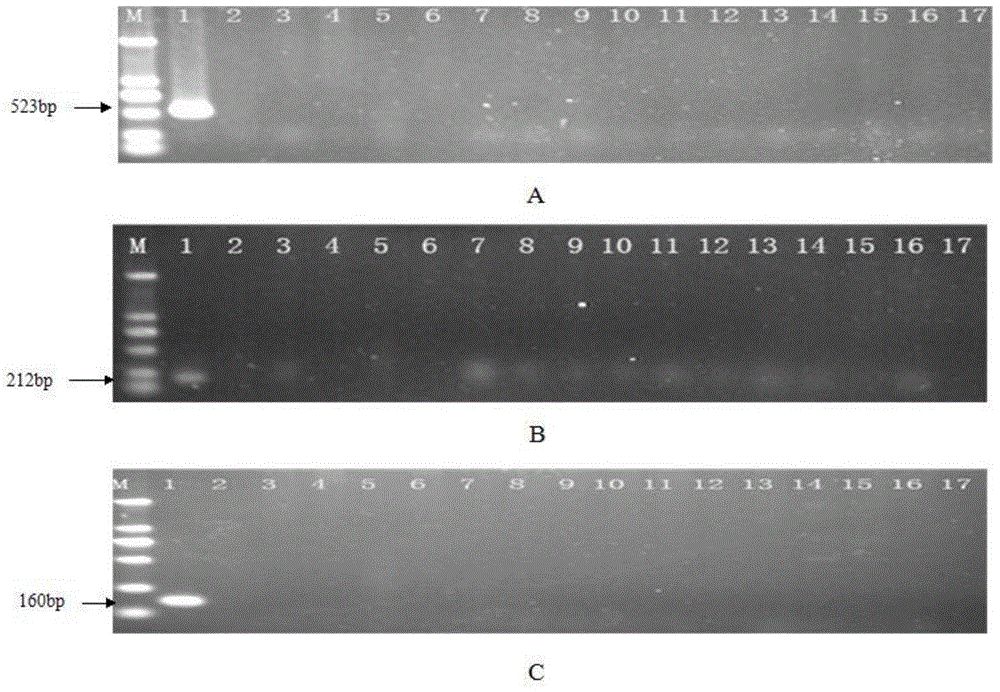
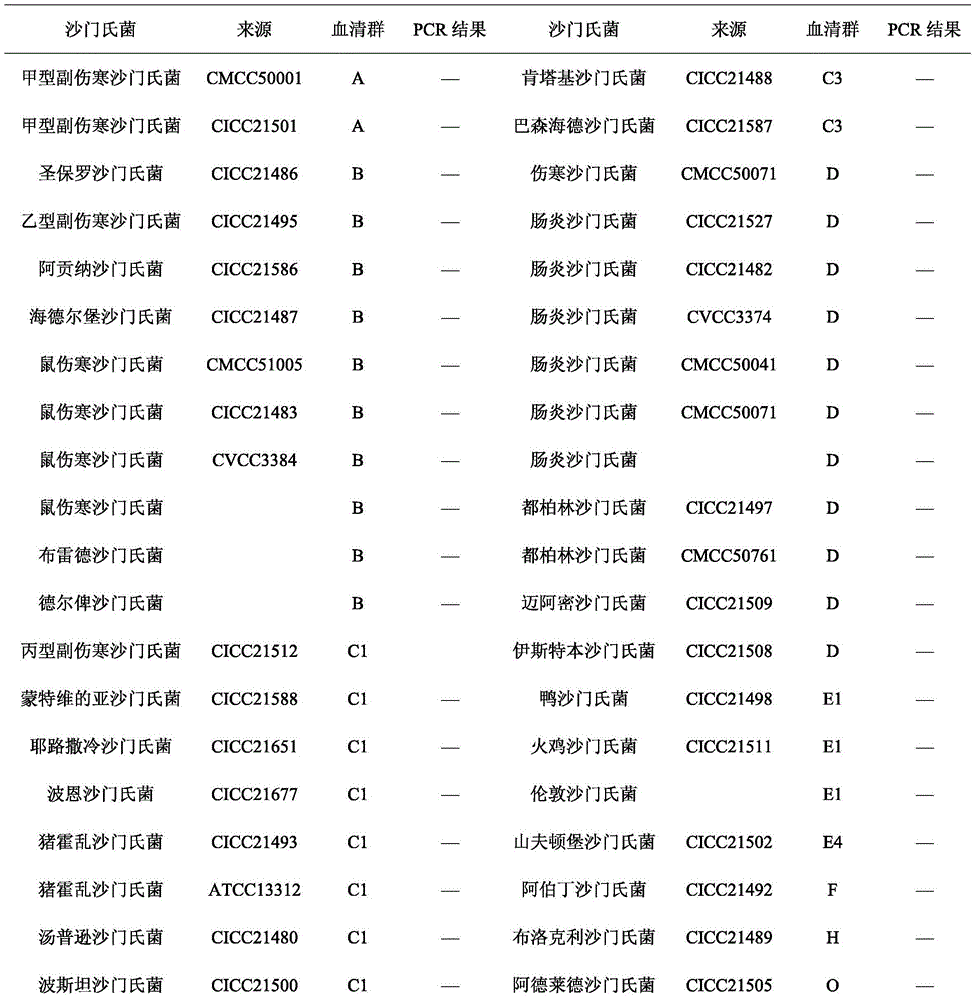
[0032] Specificity evaluation of different bacterial species by PCR detection method
[0033] 1. Screening of serotype-specific genes of Salmonella paratyphi C
[0034] The complete genome sequence of Salmonella paratyphi C RKS4594 strain (NC_012125.1) was obtained in the NCBI database. According to the method of comparing genomes, each gene of the genome of the bacteria was compared using the Blast program of the NCBA website, and the serum was selected. The genes with high type homology (E value=0, Query cover=0%) and low homology with other microorganisms (Query cover<10%) were used as quasi-specific genes of Salmonella paratyphi C. A total of 7 quasi-specific genes of Salmonella paratyphi C were obtained by this method, and multiple pairs of primers were designed for each gene, and the specificity was verified by using other serotypes of Salmonella preserved in the laboratory. According to the PCR results, the SPC_0871, SPC_0872 and SPC_0908 genes were determined as the s...
Embodiment 2
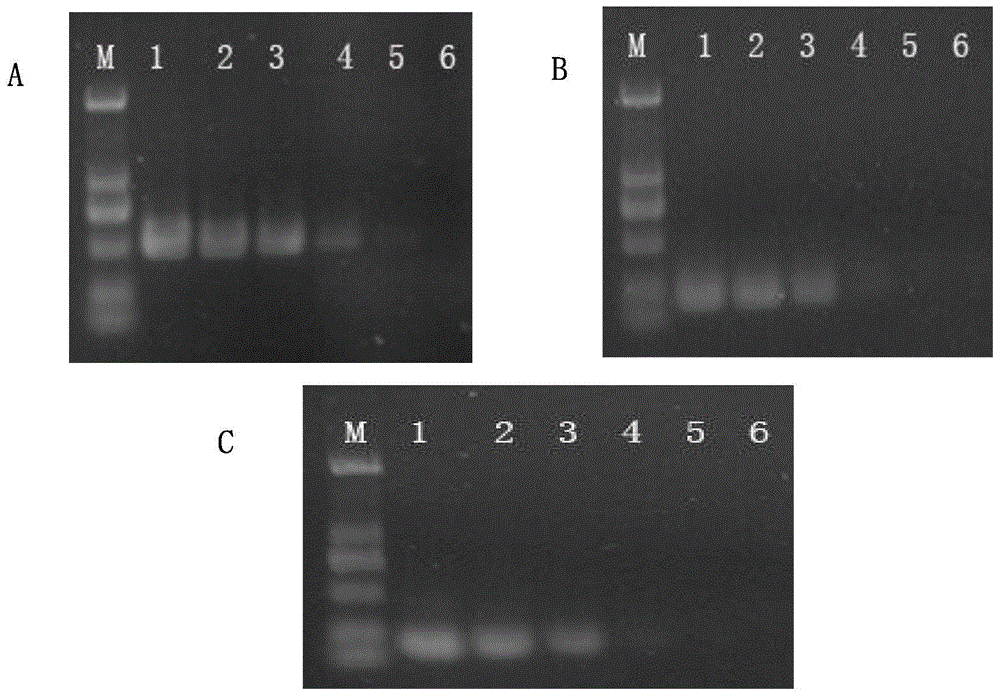
[0047] Sensitivity Evaluation of PCR Detection Method to Different Template Concentrations
[0048] The screening and primer design steps of Salmonella paratyphi C serotype-specific genes are as in Example 1.
[0049] Preparation of DNA template: Pick a single colony of Salmonella paratyphi C and transfer it to 30 ml of LB liquid medium, and cultivate overnight at 37°C. The bacterial genomic DNA mini-extraction kit produced by Shanghai Sangon Bioengineering Technology Service Co., Ltd. was used to extract the total DNA of Salmonella paratyphi C, and the concentration was determined to be 98.33ng / ul. Make a 10-fold gradient dilution with sterile water, and dilute a total of 6 gradients as a PCR template.
[0050] Establishment of PCR reaction system: establishment of PCR detection system: 25ul reaction system includes: 2×Master 12.5ul, 5uM primer pair 1ul, template 5ul, supplemented with ddH 2 0 to 25ul. The reaction program of PCR was designed: pre-denaturation at 94°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com