Apolygus lucorum vitellogenin and specific peptide chain, vector, strain and application thereof
A technology of vitellogenin and chlorophyll, which is applied in the field of agricultural science and can solve the problems of lack of detection methods for protein expression and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1: the acquisition of AlVg full-length gene
[0044]The TRIzol method was used to extract the total RNA of Lygus japonica, and the total RNA samples with good electrophoretic patterns and OD260 / OD280 values between 1.8 and 2.0 were selected and combined for mRNA purification, and the first-strand cDNA was synthesized by reverse transcription. The PCR reaction system was: 0.5 μg total RNA, 0.5μl Oligo-dT18, 2μl 5× reaction buffer, 2μl 10mM dNTP, 40U RNasin, 200U SuperscriptⅡ, add ddH 2 0 to 10 μl; PCR reaction parameters: 42°C for 30 min, 75°C for 30 min, 4°C for 5 min. According to the known conserved sequence of insect Vg gene, primers AlVg-1F (5'-CGTTGGTAGCCGCATGAAC-3') and AlVg-1R (5'-GAACTGCTGCTGGAACTGCTG-3') suitable for PCR amplification were designed to obtain the conserved sequence of AlVg gene. For the sequence, refer to SEQ ID NO: 19 in the sequence listing. The PCR reaction system is: 2.5 μl 10× reaction buffer, 2 μl 25 mM Mg 2+ , 2 μl 10 mM dN...
Embodiment 2
[0046] Embodiment 2: AlVg protein-specific peptide chain cDNA clone
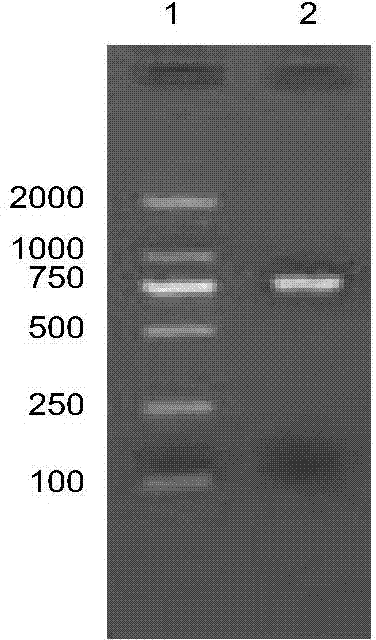
[0047] According to the full-length sequence of the AlVg gene, the cDNA sequence of the specific peptide chain of the AlVg protein is predicted in the national authoritative gene database National Center for Biotechnology Information (NCBI). The base is shown in SEQ ID NO: 3, the amino acid sequence is shown in SEQ ID NO: 4, and Design PCR amplification primers AlVg-3F (5′-CATATGCAAATCCAAATCTCAACGT-3′) and AlVg-3R (5′-TCTAGATCAAGCTTTGCAGCGAG-3′) to obtain AlVg protein specific peptide chain gene sequence, PCR reaction system: 2.5μl 10× Reaction buffer, 2 μl 25mM Mg 2+ , 2 μl 10 mM dNTP, 0.5 μl Taq plus DNA polymerase, 1 μl 10 μM upstream primer AlVg-1-F, 1 μl 10 μM downstream primer AlVg-1-R, 1 μl cDNA template, add ddH 2 O to 25 μl; PCR reaction parameters: 94°C for 5 minutes; 94°C for 30s, 60.5°C for 30s, 72°C for 60s, 40 cycles; 72°C for 10 minutes. The results of electrophoresis showed that after PCR a...
Embodiment 3
[0049] Embodiment 3: Construction of AlVg protein-specific peptide chain DNA prokaryotic expression vector
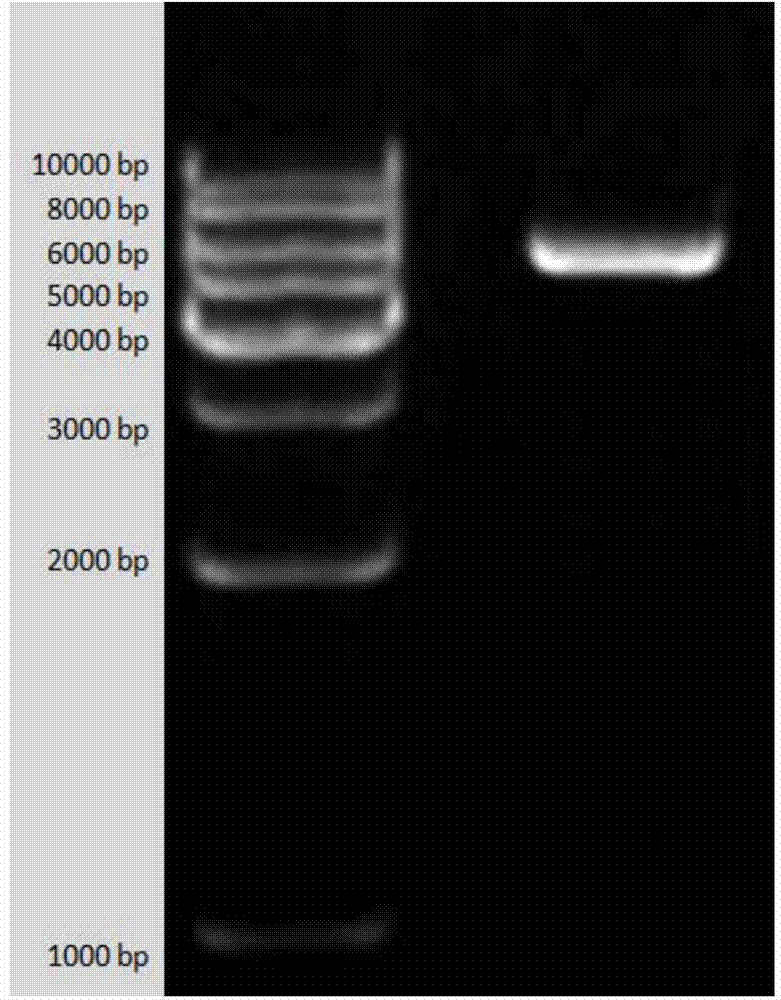
[0050] The method for constructing the prokaryotic expression vector containing the AlVg protein-specific peptide chain gene T vector is as follows: the above-mentioned sequencing is correctly connected into the pGEM-TEasy vector and pCzn1 are both digested with restriction endonucleases NdeI and XbaI, and connected. The enzyme digestion system is: 1.0 μl of each of the two restriction enzymes, 2.0 μl of 10×Buffer buffer, 10.0 μl of gene fragments with appropriate restriction sites, and ddH 2 Make up O to 20 μl, and bathe in water at 37°C for 3h. The ligation system is: 5.0 μl of the recovered vector after enzyme digestion, 10.0 μl of the gene fragment, 1.0 μl of T4 DNA ligase, 2.0 μl of 10×Buffer, and ddH 2 Make up O to 20 μl, react at 16°C for 12-16 hours, and see the enzyme digestion map after successful ligation Figure 4 . After the ligation product was transfor...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com