Spodoptera exigua vitellogenin receptor, specific peptide chain, vector, strain and application thereof
A technology of vitellogenin, beet armyworm, applied in the field of agricultural science
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1: Obtaining of SEVgR full-length gene
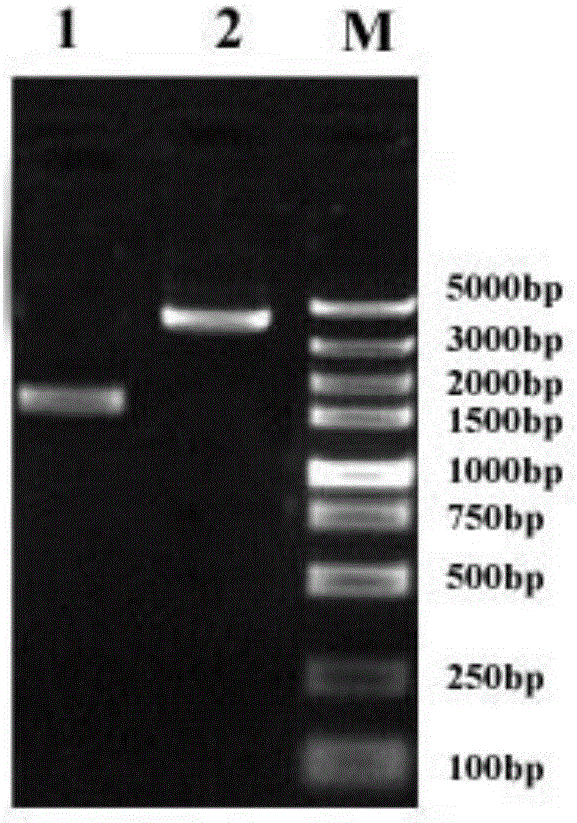
[0037] The total RNA of beet armyworm was extracted by TRIzol method, and the total RNA samples with good electrophoretic pattern and OD260 / OD280 value between 1.8 and 2.0 were selected and combined for mRNA purification, and the first-strand cDNA was synthesized by reverse transcription. The PCR reaction system was: 0.5 μg total RNA, 2.5μl Oligo-dT18, 5μl MMlv Buffer, 5μl 10mM dNTP, 40U RNasin, 1μl MMlv reverse transcriptase (reverse transcriptase), add ddH 2 O to 25 μl; PCR reaction parameters: 42°C for 1h, 95°C for 10min. According to the conserved sequence of the known insect VgR gene, primers SEVgR-3396F (5'-CGAAGAGGACTGTGATGAACCT-3') and SEVgR-4845R (5'-GACAAATGTGTTGGCGTCAA-3') suitable for PCR amplification were designed to obtain the conserved sequence of the SEVgR gene. For the sequence, refer to SEQ ID NO: 11 in the sequence listing, the PCR reaction system is: 5 μl 10× reaction buffer, 5 μl 25mM Mg 2+, 8μl...
Embodiment 2
[0039] Embodiment 2: SEVgR specific peptide chain cDNA clone
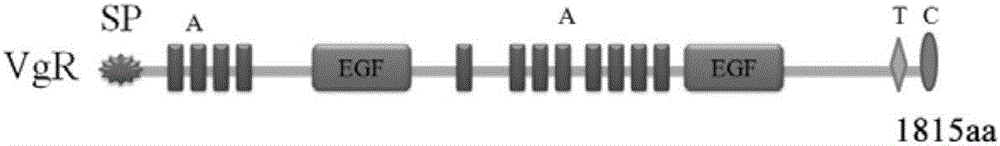
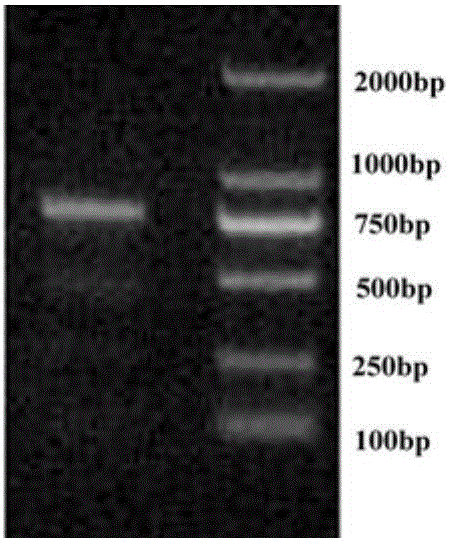
[0040] According to the measured full-length sequence of the SEVgR gene, DNAMAN software was used to predict the cDNA sequence of the SEVgR specific peptide chain, and PCR amplification primers SEVgR-3F (5′-TGTCATGAAACTGAGTTCATGTG-3′) and SEVgR-3R (5′-GCATGAGAACTGATCCTTGGACG- 3') to obtain the SEVgR-specific peptide chain gene sequence, the PCR reaction system is: 5 μl 10× reaction buffer, 5 μl 25mM Mg 2+ , 8 μl 10 mM dNTP, 0.5 μl Takara LA DNA polymerase, 1 μl 10 μM upstream primer SEVgR-3F, 1 μl 10 μM downstream primer SEVgR-3R, 1 μl cDNA template prepared in Example 1, add ddH 2 O to 50 μl; PCR reaction parameters: 94°C for 5 minutes; 35 cycles of 94°C for 30s, 55°C for 30s, 72°C for 1 min; 72°C for 10 minutes. The results of electrophoresis showed that after PCR amplification and splicing, a band consistent with the expected size of 786bp was obtained ( image 3 ). The PCR product was separated by 1.2% aga...
Embodiment 3
[0042] Example 3: Construction of SEVgR-specific peptide chain DNA prokaryotic expression vector
[0043] The method for constructing the prokaryotic expression vector containing the SEVgR-specific peptide chain gene T vector is as follows: link the correctly sequenced PCR product of Example 2 into the expression vector pET15B through the cloning sites Nde I and Xho I, and verify whether a recombinant plasmid is formed by enzyme digestion. The digestion system is: 0.25 μl each of the two restriction enzymes (NdeI and XbaI), 1.0 μl of 10×Buffer buffer, 3 μl of plasmid, and ddH 2 Make up O to 10 μl, and bathe in water at 37°C for 3h. For the enzyme digestion map after successful connection, see Figure 4 . Transform the correctly identified recombinant plasmid named pET15B-SEVgR into 100 μl Escherichia coli Arctic Express (Zhongding Biological Company) competent cells, put it on ice for 20 minutes, heat shock at 42°C for 90 seconds, quickly put it in ice for 5 minutes, and add...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com