Method for controlling and improving DNAzyme catalytic activity
A technology with high catalytic activity and catalytic activity, applied in the field of biological analysis, can solve the problem of limited catalytic activity of deoxyribozyme
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
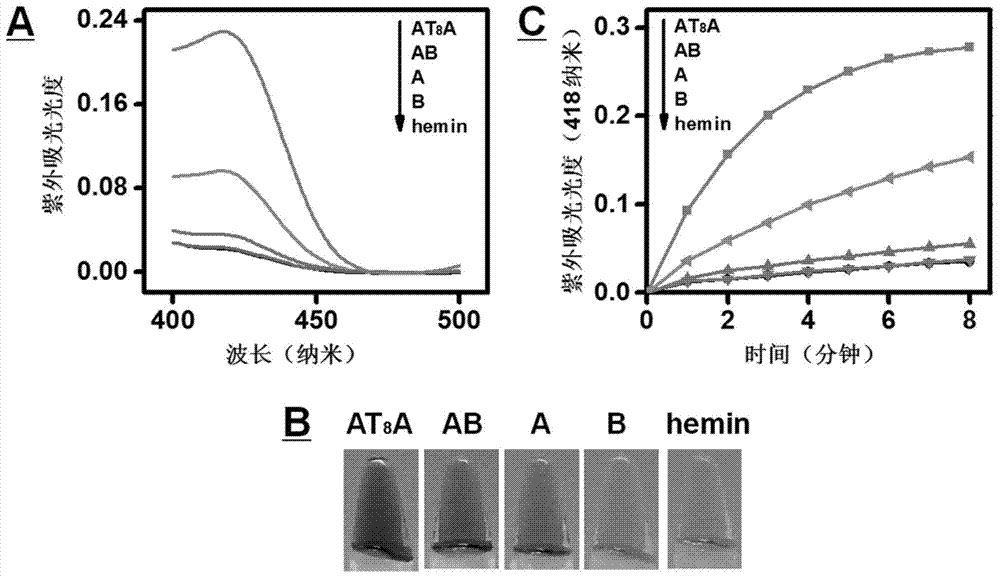
[0017] Embodiment 1: four deoxyribozymes (A, B, AB and AT 8 A) Catalytic hydrogen peroxide oxidation of ABTS by -hemin complex 2+ comparison of reactivity.
[0018] Four ribozymes (A, B, AB and AT 8 A, the sequence is shown in Table 1) dissolved in reaction buffer I (4-hydroxyethylpiperazineethanesulfonic acid (HEPES) (25mM), KNO 3 (20mM),NaNO 3 (200mM), Triton X-100 (0.025% (w / v)), DMSO (1% (v / v)), pH 5.3), the final concentration is 0.125μM. The above solution was heated in a 95℃ water bath 6 minutes, then slowly cooled to room temperature (about 25° C.) for 2 hours. Hemin (final concentration: 0.2 μM) was added to the heat-treated solution, and incubated at room temperature for 1 hour to form a deoxyribozyme-hemin complex. Add reactant ABTS 2+ (final concentration 0.5mM) and H 2 o 2 (final concentration 1mM) in the above solution. Record the green product ABTS when the reaction proceeds for 4 minutes ·+ UV absorption spectrum. The maximum absorbance value of the ...
Embodiment 2
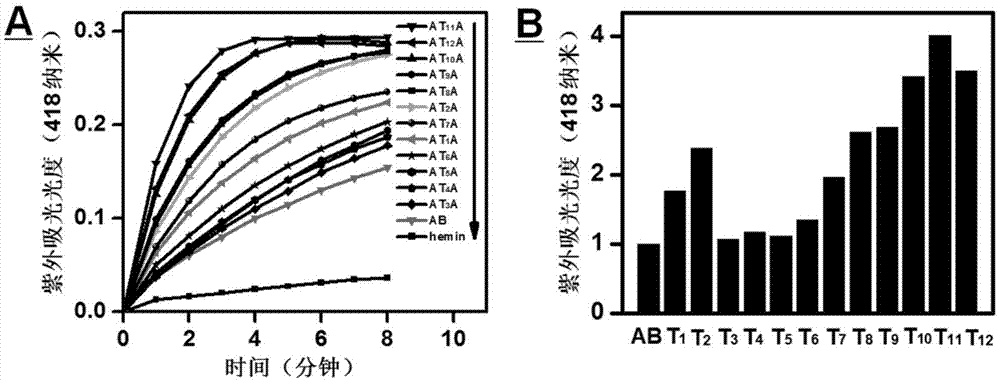
[0020] Embodiment 2: four kinds of deoxyribozymes (A, B, AB and AT 8 A) - Comparison with kinetic data for hemin-catalyzed peroxide reactions.
[0021] Four ribozymes (A, B, AB, and AT 8 A, the sequence is shown in Table 1) dissolved in reaction buffer I (HEPES (25mM), KNO 3 (20mM),NaNO 3 (200 mM), Triton X-100 (0.025% (w / v)), DMSO (1% (v / v)), pH=5.3), the final concentration was 0.125 μM. The solution was first heated in a 95°C water bath for 6 minutes, and then slowly cooled to room temperature (about 25°C) for 2 hours. Hemin (hemin) with a final concentration of 0.2 μM was added to the above heat-treated solution, and co-incubated at room temperature for 1 hour to form a deoxyribozyme-hemin complex. Add reactant ABTS 2+ (final concentration 0.5mM) and H 2 o 2 (final concentration 1mM) in the above solution. Record AB-hemin, A-hemin, B-hemin, AT every 1 minute 8 A-hemin and hemin itself catalyze H 2 o 2 Oxidized ABTS 2+ The maximum ultraviolet absorbance value (a...
Embodiment 3
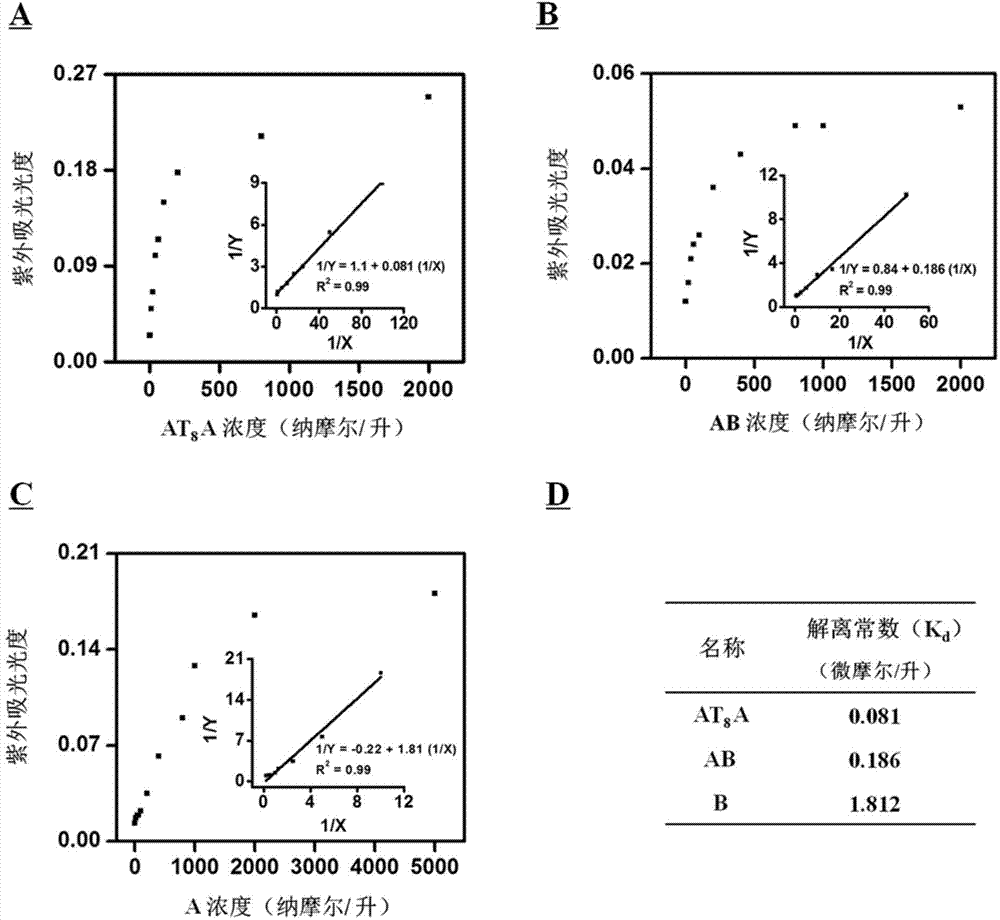
[0023] Embodiment 3: Three kinds of deoxyribozyme dissociation constants K d measurement and comparison.
[0024] Three deoxyribozymes AT 8 A, AB and A were respectively dissolved in reaction buffer I, and the final concentration ranges were OnM, 10 nM, 20 nM, 40 nM, 60 nM, 100 nM, 200 nM, 800 nM, 2 μM to 5 μM. The solutions were first heated in a water bath at 95° C. for 6 minutes, and then cooled slowly to room temperature (about 25° C.) for 2 hours. Hemin (hemin) with a final concentration of 0.2 μM was added to the above heat-treated DNA solution, and co-incubated at room temperature for 1 hour to form a deoxyribozyme-hemin complex. Add reactant ABTS 2+ (final concentration 0.5mM) and H 2 o 2 (final concentration 1mM) in the above solution. Record the UV absorbance value of the reaction mixture at different concentrations at 1 minute.
[0025] In the inset graph, X represents the concentration of DNAzyme (in μM). The calculation formula of Y is, Y=(A x -A 0 ) / (A ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com