DNA flexagon nano structure-nanogold biosensor based on adapter modification and preparing method and application of DNA flexagon nano structure-nanogold biosensor
A biosensor and nanostructure technology, applied in the field of detection and analysis, to achieve the effects of reducing costs, overcoming poor autoimmunity, and low detection limits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1 Preparation of equilateral triangle DNA origami
[0032] According to the sequence designed by Rothemund (P.W.Rothemund, Nature, 2006, 440, 297.0), the M13mp18 phage (purchased from NEB Company, item number: #N4040S), more than 200 unmodified staple chains (P.W. Rothemund, Nature, 2006, 440, 297.0 ), the staple chain (SEQ ID No.1) modified with aflatoxin aptamer at the end was mixed according to the molar ratio of 1:10:10, and then 1×TAE~Mg 2+ Buffer (Mg 2+ After the concentration of 12.5mM), the mixed solution was placed in a PCR instrument and annealed from 95°C to 20°C at a rate of 1°C / 100s. After the reaction, a 100kDa ultrafiltration tube (Amicon Ultra Company) was used to remove excess staple chains and then put into Next use.
[0033] Wherein, the staple chain sequence with the aflatoxin aptamer modified at the end is as follows:
[0034] 5'-gttgggcacg tgttgtctct ctgtgtctcg tgcccttcgc taggcccact ttttgtgaga aaatgtgtag gtaaagatac aacttt-3', (SEQ ID ...
Embodiment 2
[0035] Example 2 Preparation of SH-DNA modified gold nanoparticles
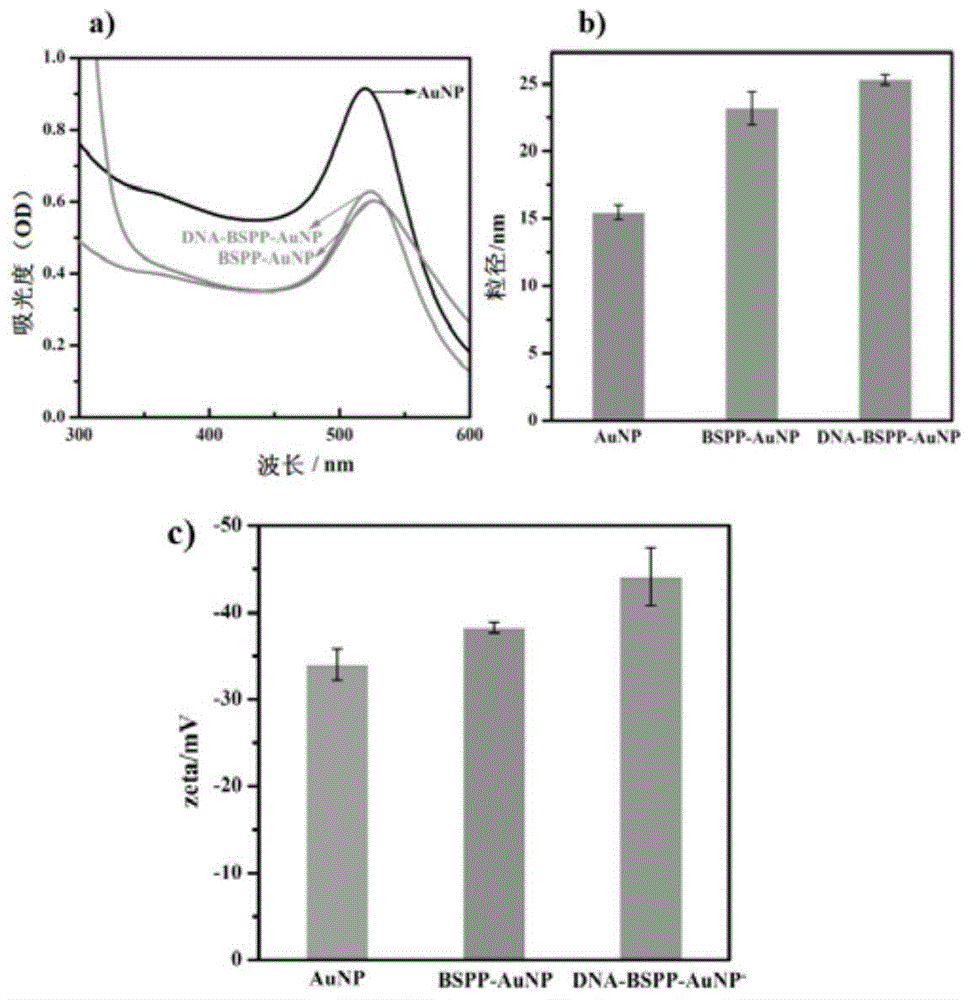
[0036] BSPP (bis(p-sulfonatophenyl)phenylphosphine dihydrate dipotassium salt, purchased from Sigma-Aldrich Company) was used to modify gold nanoparticles, using the method reported by Ding Baoquan (B.Q.Ding, Z.T.Deng, H.Yan, S.Cabrini, R.N.Zuckermann, J. Bokor, J.AM.CHEM.SOC, 2010, 132, 3248): 15mg BSPP was added to 50mL nano-gold solution with a concentration of 1.67nM, and after shaking at room temperature for 12h in the dark, solid sodium chloride was added until the solution color changed to Light purple, centrifuge at 10000rpm for 15min, discard the supernatant, resuspend the pellet in 1mL of 2.5mM BSPP solution, add 1mL of methanol, and centrifuge again, collect the pellet, and resuspend in BSPP. The UV-Vis spectrophotometer, particle size, and zeta potential characterization results before and after nano-gold modification are shown in figure 1 , it can be seen that after each step of modification, t...
Embodiment 3
[0040] Example 3 Feasibility experiment of aflatoxin B1 detection based on aptamer-modified DNA origami nanostructure-nanogold biosensor
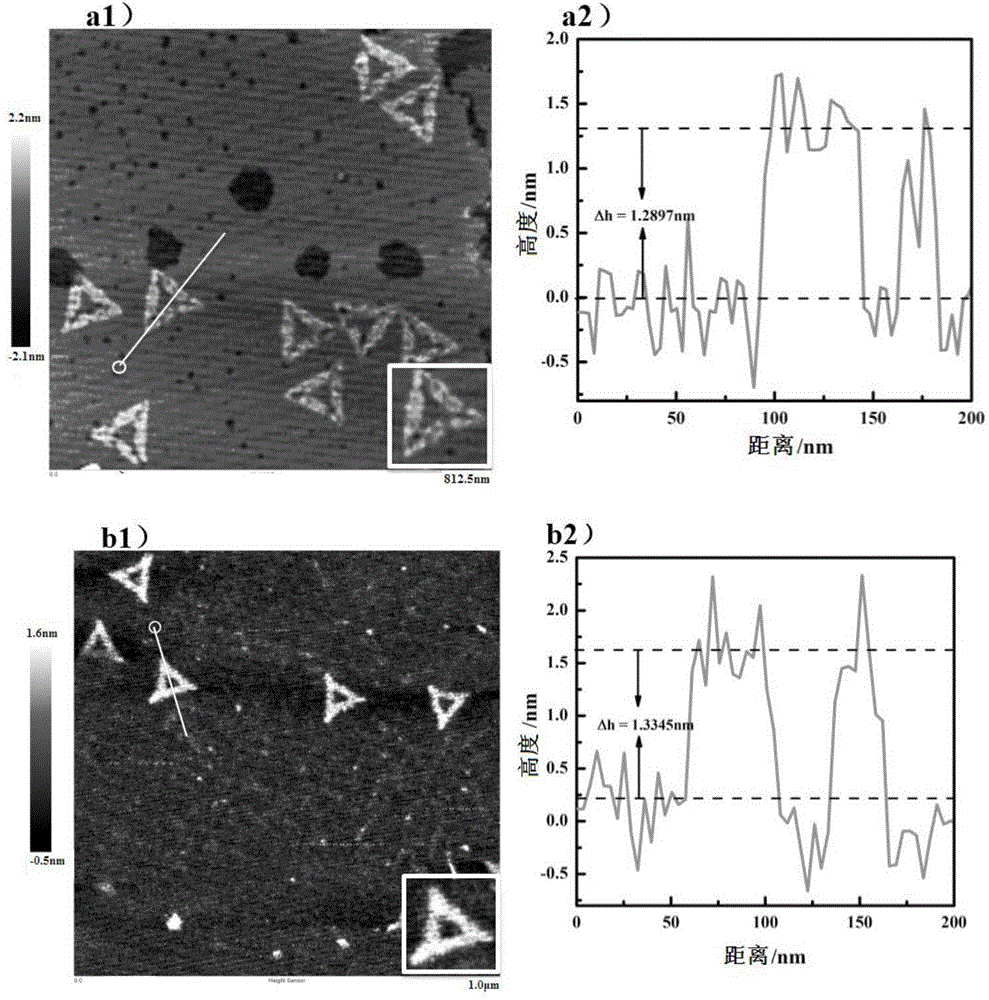
[0041] Firstly, the DNA origami with the aflatoxin aptamer is specifically reacted with the aflatoxin: the DNA origami with the aflatoxin aptamer is mixed with excess aflatoxin at a molar ratio of 1:100 , react at room temperature for 30 minutes, take the newly dissociated mica flakes, drop into 10 μL of the above-mentioned mixture after the reaction, and use the ScanAsyst mode to perform characterization with an atomic force microscope (Bruker DIMENSION icon) after drying. The characterization results are as follows figure 2 It is shown in figure b1 in middle.
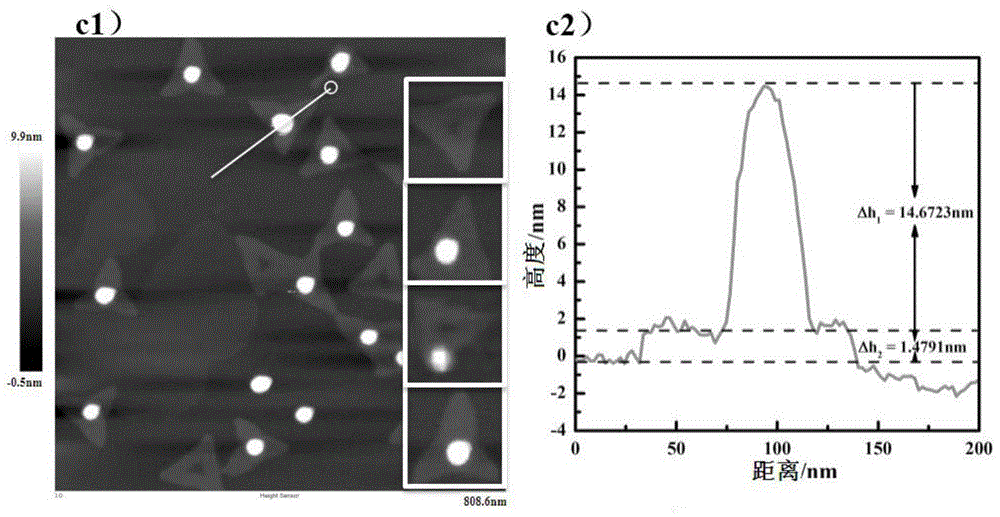
[0042] Then hybridize the DNA origami connected with the aflatoxin aptamer and the gold nanoparticles modified with complementary DNA at a molar ratio of 1:5, put it into a PCR instrument for gradient annealing at 43°C-20°C, and take a new solution 10 μL of the reacted above-ment...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com