Method and kit for rapid construction of single-cell DNA sequencing library
A DNA sequencing, single-cell technology, applied in the field of single-cell DNA sequencing library construction, can solve the problems of complex process, unsuitable for large-scale production applications, poor genome coverage and uniformity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
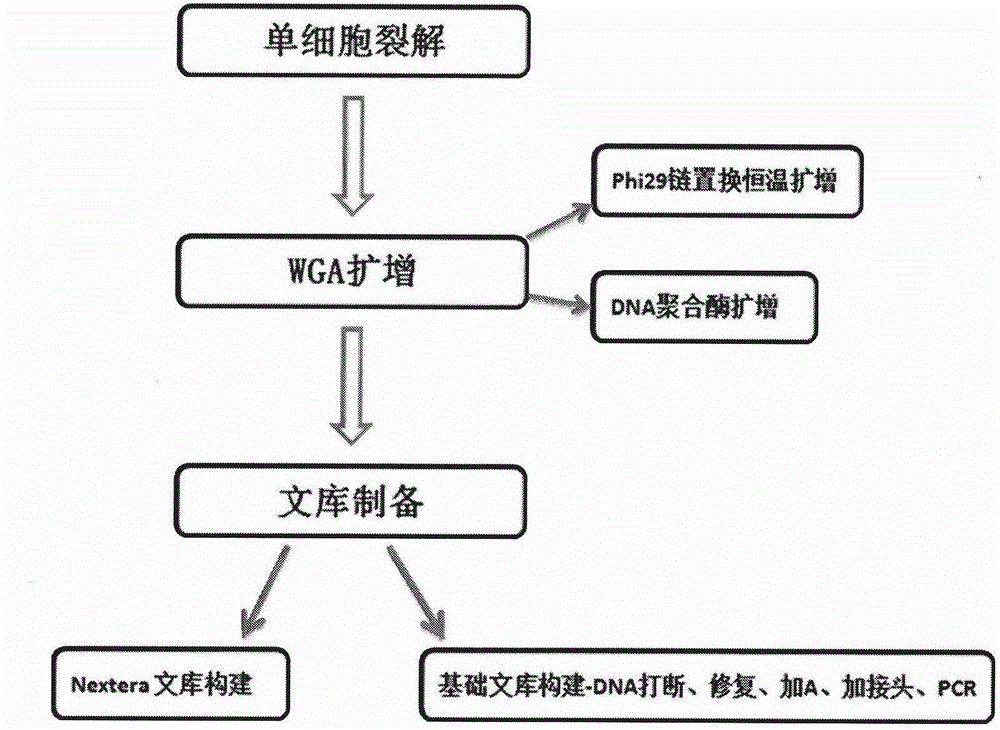
[0078] The construction method of the single-cell DNA sequencing library of Example 1 mainly includes the following steps (see image 3 ):
[0079] (1) Cell lysis: This step can be performed using any protease, protein denaturing reagent and lysis buffer system known to those skilled in the art that are suitable for cell lysis.
[0080] (2) Pre-amplification of genomic DNA can be performed using any thermostable DNA polymerase known to those skilled in the art, for example: LA-Taq, rTaq, Phusion, Deep Vent, Deep Vent (exo-), Gold 360 , Platinum Taq, KAPA 2G Robust.
[0081] The preamplification primers used in the present invention are mainly composed of three parts: a common region, which has different sequences in different sequencing platforms; a degenerate base region, which only contains two kinds of bases, A and C, or A and G , or T and C, or T and G; a region of random degenerate bases, where each base can be A, T, C, or G, and two sulfurs are added between the last t...
Embodiment 2
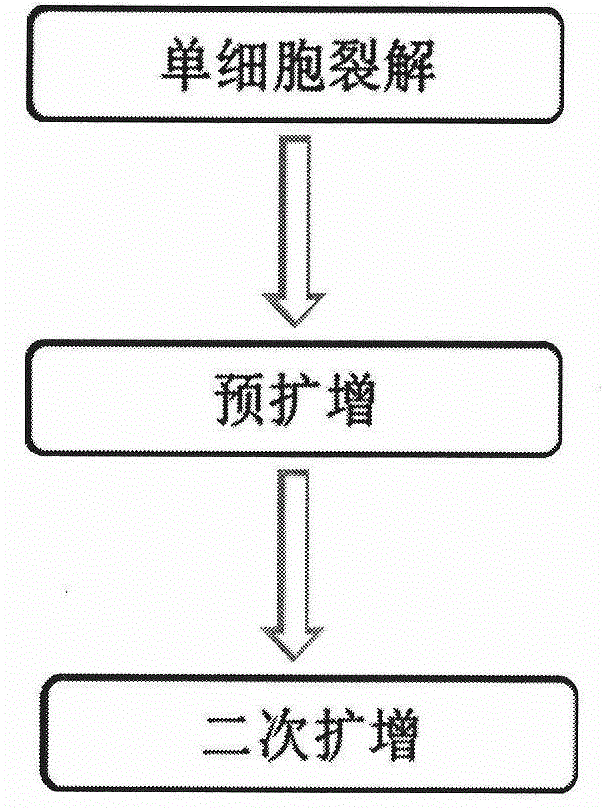
[0114] The library construction method in Example 2 is basically the same as in Example 1, the difference is that in Example 2, the starting sample is multi-cell rather than single-cell (please refer to image 3 ).
[0115] A specific example of performing multicellular DNA sample sequencing according to the method of Embodiment 2 of the present invention is shown below
[0116] Step 1: Multiple cell lysis. Prepare cell lysis buffer as shown in Table 1, incubate at 60°C for 20 minutes, 95°C for 4 minutes, then keep samples at 4°C.
[0117] Table 1
[0118]
[0119] Step 2: Genomic DNA pre-amplification. Prepare the pre-amplification PCR system shown in Table 2.
[0120] Table 2
[0121]
[0122] The PCR reaction scheme is: a.95°C for 3 minutes→b.16 cycles as follows: 98°C for 20 seconds, 15°C for 50 seconds, 25°C for 40 seconds, 35°C for 30 seconds, 65°C for 40 seconds, 72°C for 1 minute→ c. Keep at 4°C.
[0123] Step 3: Secondary amplification of genomic DNA. Pr...
Embodiment 3
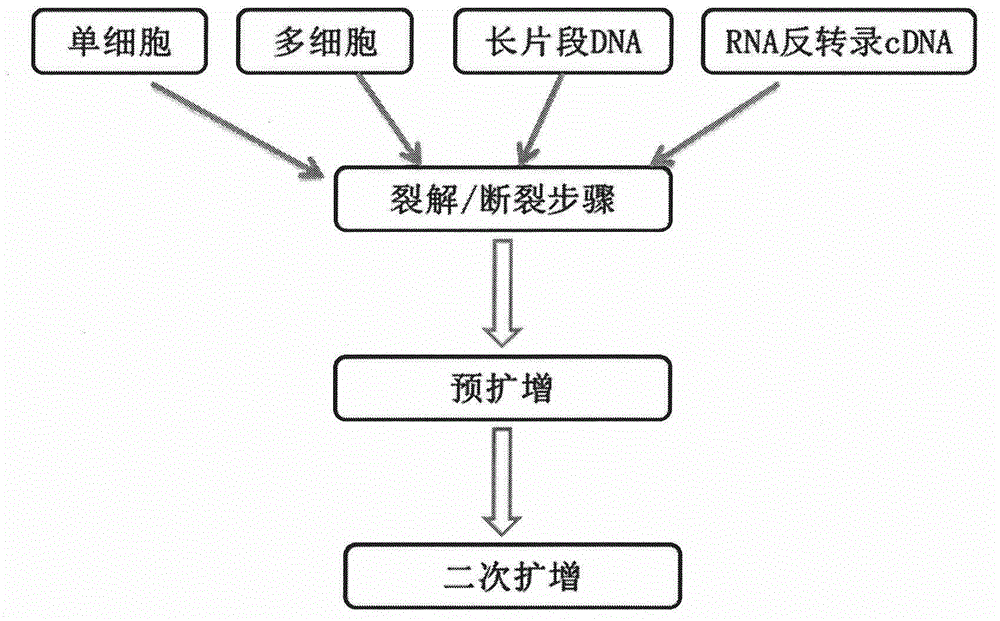
[0132] The library construction method in Example 3 is basically the same as in Example 1, except that in Example 3, the starting sample is DNA instead of cells. In this example, the starting sample can be genomic DNA, mitochondrial DNA, long PCR products, or long chromatin-precipitated DNA (see image 3 ).
[0133] A specific example of performing DNA sample sequencing according to the method of Embodiment 3 of the present invention is shown below
[0134] Step 1: Fragmentation of long fragments of DNA. Prepare cell lysis buffer as shown in Table 1, incubate at 60°C for 20 minutes, 95°C for 4 minutes, then keep samples at 4°C. Alternatively, DNA fragments were directly incubated at 95°C for 4 minutes, and then the samples were kept at 4°C.
[0135] Table 1
[0136]
[0137] Step 2: DNA pre-amplification. Prepare the pre-amplification PCR system shown in Table 2.
[0138] Table 2
[0139]
[0140] The PCR reaction scheme is: a.95°C for 3 minutes→b.16 cycles as follo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com