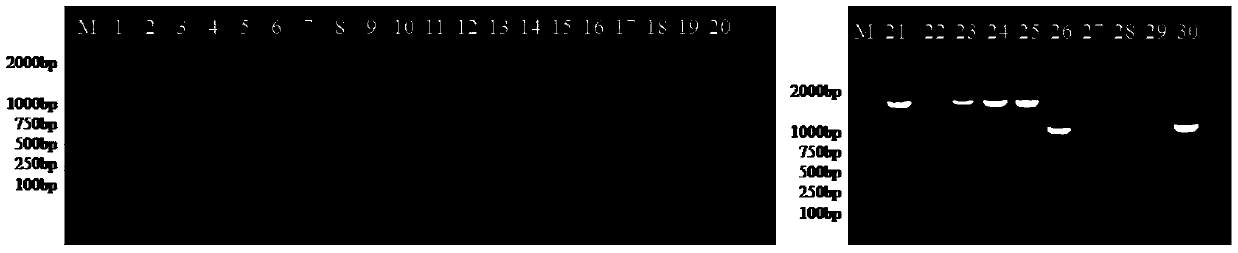A kind of gii.4 type norovirus genome amplification primer and amplification method
A virus genome and amplification primer technology, which is applied in the field of GII.4 norovirus genome amplification primers and amplification, can solve the problems of difficult sensitivity, large fragment length, and increased difficulty of amplification of direct amplification methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Optimization of RT-PCR annealing temperature for primers suitable for GII.4 norovirus genome amplification
[0041] (1) Virus sample processing and nucleic acid extraction: Dilute the collected sample to be processed (containing GII.4 Norovirus L10) to a concentration of 10% (w / v) with PBS solution (pH7.4, treated with DEPC), shake fully Mix well, and centrifuge at 12000×g for 10 min to collect 140 μL of the supernatant, and extract 60 μL of viral RNA in the sample with an RNA extraction kit.
[0042](2) "4+1+1" genome segment amplification method (that is, amplification in 6 segments, the primers used are as follows: P1F / P1596R, P1258F / P2805R, P2689F / P4248R, P4099F / G2SKR, NV2of2 / GV132 and P6484F / P7513R, select a pair of primers for each RT-PCR reaction): use 20 μL of one-step RT-PCR reaction system, containing 10 μL of 2×one-stepRT-PCR mixture, upstream primer and downstream primer (10 μmol / L) 0.6 μL each, 0.8 μL of MLV / RNasin / HS-Taq enzyme mixture, 2 μL of sample vi...
Embodiment 2
[0046] Embodiment 2: RT-PCR sensitivity analysis applicable to primers for GII.4 type norovirus genome amplification
[0047] (1) Virus sample processing and nucleic acid extraction: Dilute the collected sample to be processed (containing GII.4 Norovirus L10) to a concentration of 10% (w / v) with PBS solution (pH7.4, treated with DEPC), shake fully Mix evenly, collect 140 μL of supernatant by centrifugation at 12000×g for 10 min, and extract 60 μL of viral RNA in the sample with an RNA extraction kit, and perform appropriate dilution of 10× gradient.
[0048] (2) "4+1+1" genome segment amplification method (that is, amplification in 6 segments, the primers used are as follows: P1F / P1596R, P1258F / P2805R, P2689F / P4248R, P4099F / G2SKR, NV2of2 / GV132 and P6484F / P7513R, select a pair of primers for each RT-PCR reaction): Use 20 μL of one-step RT-PCR reaction system, containing 10 μL of 2×one-stepRT-PCR mixture, upstream primer and downstream primer (10 μmol / L) 0.6 μL each, 0.8 μL of...
Embodiment 3
[0052] Embodiment 3: Amplification effect of virus genome in actual sample
[0053] (1) Virus sample processing and nucleic acid extraction: take GII.4 norovirus positive samples L10, L62, L65, L106, L232, and dilute the sample to be treated to 10% (w) by PBS solution (pH7.4, DEPC treatment) / v) concentration, shake and mix well, centrifuge at 12000×g for 10 min to collect 140 μL of supernatant, and extract 60 μL of viral RNA in the sample by RNA extraction kit.
[0054] (2) "4+1+1" genome segment amplification method (that is, amplification in 6 segments, the primers used are as follows: P1F / P1596R, P1258F / P2805R, P2689F / P4248R, P4099F / G2SKR, NV2of2 / GV132 and P6484F / P7513R, select a pair of primers for each RT-PCR reaction): Use 20 μL of one-step RT-PCR reaction system, containing 10 μL of 2×one-stepRT-PCR mixture, upstream primer and downstream primer (10 μmol / L) 0.6 μL each, 0.8 μL of MLV / RNasin / HS-Taq enzyme mixture, 2 μL of sample RNA template, and the rest by ddH 2 O ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com