A method for identifying high-yield traits of royal jelly in bee colonies using the snp marker rs4208349
A bee colony royal jelly, high-yield technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of high cost, complex identification process, unfavorable popularization, etc., to improve the success rate, rapid identification, and efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
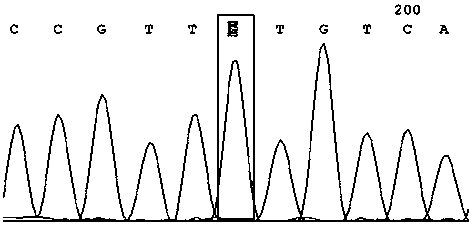
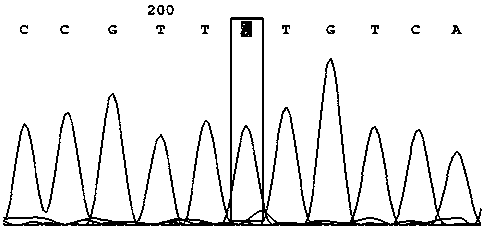
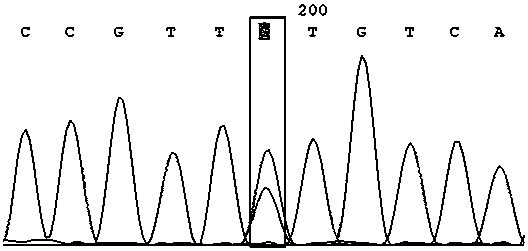
[0040] A method utilizing SNP marker rs4208349 to identify high-yield traits of honey bee colony royal jelly, comprising the following steps:
[0041] (1) Sampling of bees: 50 adult worker bee individuals were collected from the bee species with high royal jelly production ("Zhenongda No. 1") and the bee species with low royal jelly production (Apis mellifera, USA), and each individual was placed in a centrifuge tube. Freeze at -20°C for later use;
[0042] (2) Extraction of DNA from bee samples (using Sangon’s Animal Genome Rapid Extraction Kit, SK8222)
[0043] Use ophthalmic scissors to cut off the limbs, wings and abdomen of the individual worker bees in step (1), keep only the head and thorax, put them in a mortar filled with liquid nitrogen, crush them with a grinding rod, and then transfer them to a 1.5 mL centrifuge tube Add 400 μL Buffer Digestion, shake evenly, and place in a water bath at 65 °C for 1 h until the cells are completely lysed. Then remove the supern...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com