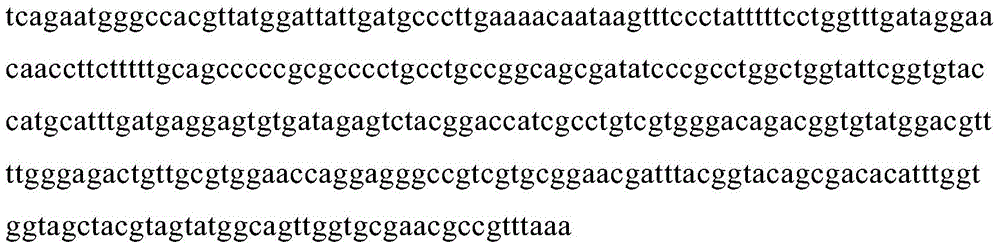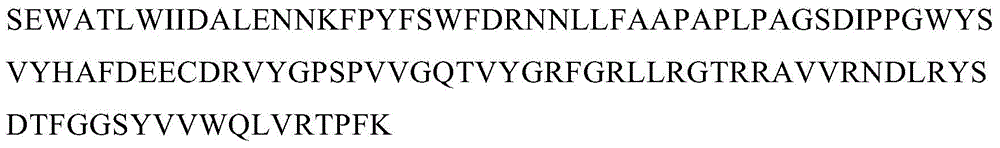KSHV virus vIRF4 DNA binding domain and polyclonal antibody thereof, and preparation method of polyclonal antibody
A technology that combines domains and viruses, applied in the fields of virology and bioengineering, can solve problems such as no resistance to Kaposi's sarcoma
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
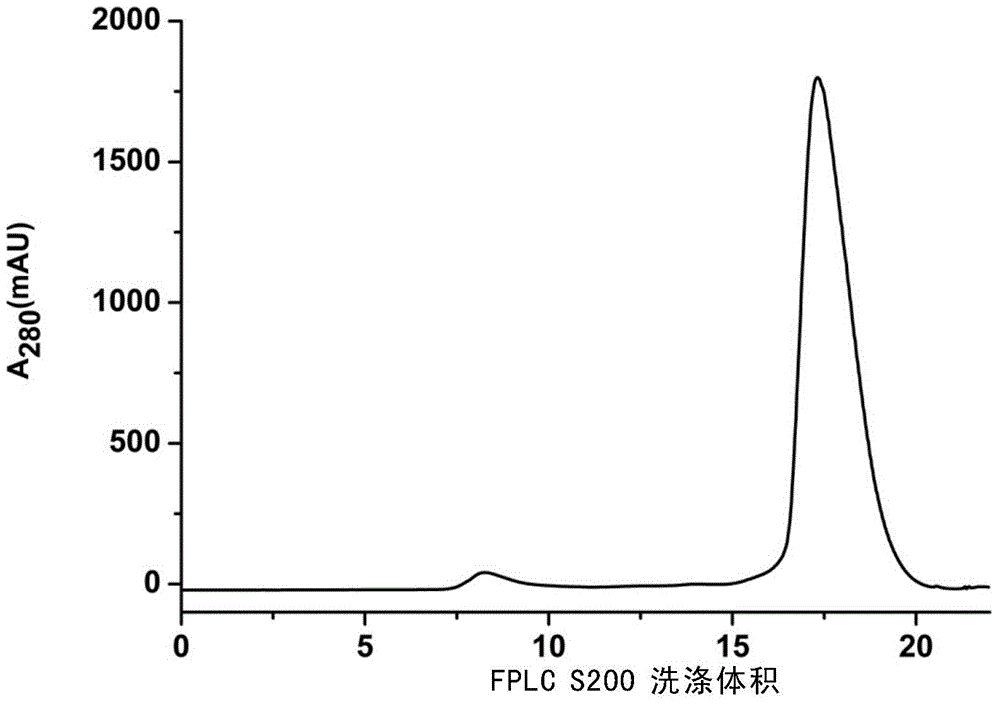
Embodiment 1
[0046] Embodiment 1, transformation of expression plasmid
[0047] pET28atp is a modified plasmid based on pET28a. The pET28 before transformation has the following sites:
[0048] NcoI-6*His-Thrombin-Nde / NheI-T7tag-BamHI / EcoRI / SacI / SalI / HindIII / NotI / XhoI-Histag;
[0049] The modified pET28a has the following sites:
[0050] NcoI-6*His-TEV-Nde / NheI-T7tag-BamHI / EcoRI / SacI / SalI / HindIII / NotI / XhoI-Histag.
[0051] It can be seen that the protease cleavage site after the 6*His tag of the plasmid before transformation is Thrombin, and the protease cleavage site after the transformation 6*His tag is TEV. There are two main reasons for the transformation based on this point: on the one hand, TEV Enzyme digestion efficiency is higher, on the other hand, the use of strains expressing TEV protease can express and purify TEV protease efficiently and rapidly.
Embodiment 2
[0052] Example 2. Obtaining of KSHVvIRF4 DNA Binding Domain Gene Fragment Sequence
[0053] Using the kshvcDNA genome as a template, design primers in Table 1 (primers were synthesized by Shanghai Sangong Company).
[0054] Table 1
[0055]
Primer sequence F cgtcgaGGATCC tcagaatgggccacgttatgg R cgtcgaCTCGAGttATTtaaacggcgttcgcaccaac
[0056] Among them, the forward primer introduces the BamHI restriction site GGATCC, the reverse primer introduces the XhoI restriction site: CTCGAG, and the base before the restriction site is the protection base. An ATT stop codon was added after the cleavage site of the reverse primer. The kit used for gene amplification comes from 2*fastpfumix of Quanshijin Company, and the PCR reaction system is shown in Table 2.
[0057] Table 2
[0058] upstream primer 2ul(20um) downstream primer 2ul(20um) genomic cDNA 20ul 2*fast pfu miX 25ul f 2 o 1ul total capacity 50ul
[...
Embodiment 3
[0062] Embodiment 3, construction contains the expression vector of target gene fragment
[0063] For the amplification of the vector pET-28a containing multiple cloning sites, the plasmid was recovered with Tiangen’s plasmid recovery kit, and the target gene fragment recovered from the gel after double-enzyme digestion and PCR amplification and the plasmid of the empty vector were respectively used. Use NEB company's BamHI restriction endonuclease and XhoI restriction endonuclease to double-digest, and the double-digestion system is shown in Table 3.
[0064] table 3
[0065] Bam H 1ul 1ul wxya 1ul 1ul CutSmart Buffer 5ul 5ul gene fragment or plasmid 30ul 30ul Add ddH 2 Total volume after O 50ul 50ul
[0066] Enzyme digestion in water bath at 37°C for 3 hours, and the target gene fragment and vector fragment were recovered by gel respectively for ligation reaction. The enzyme used to ligate the target fragment and the vector frag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com