HER2 nucleic acid aptamer and application thereof
A nucleic acid aptamer and nucleotide sequence technology, which can be used in pharmaceutical formulations, DNA/RNA fragments, biochemical equipment and methods, and can solve problems such as reducing the binding ability of nucleic acid aptamers and target molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] The preparation of embodiment 1HB3-1, HB3
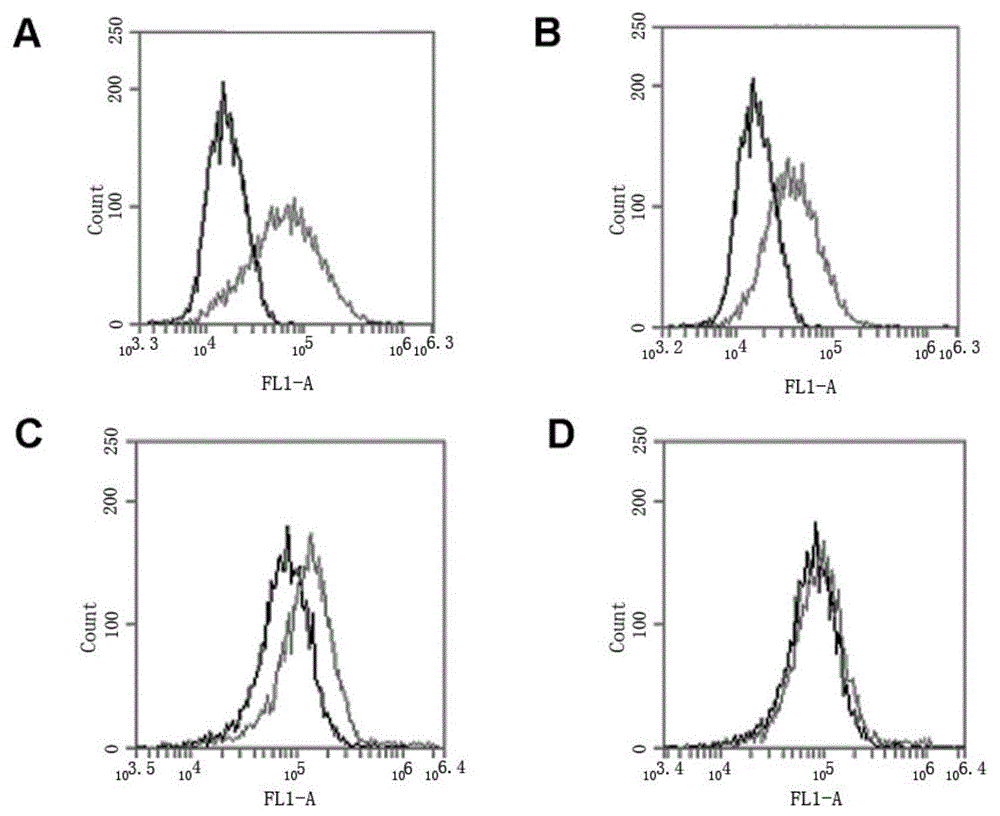
[0059] In the present invention, the core sequence of the HER2 nucleic acid aptamer is a single-stranded nucleotide with a length of 43 bases, and its nucleotide sequence is: 5'GTGTGTCTGCTATTTATTTTGCTTGATTATCTCTTAGGGATTT3'; the preferred HER2 nucleic acid aptamer is 59 bases The length of single-stranded nucleotides, the sequence is: 5'TGCCCGTGTCCCGAGGAGGTGCCCTATTTTGCTTGATTATCTCTAAGGGATTTGGGCGG-3'; more preferably, when synthesizing, all bases A in the above-mentioned sequence are modified with thio to obtain a thio HER Nucleic acid aptamers, respectively named (core sequence through thiol (HB3-1), preferred sequence through thiol (HB3)) The above sequences were all synthesized by Invitrogen. The structure of the HB3 aptamer is as follows figure 1 As shown, where A in the figure is the primary sequence, and B is the secondary structure.
[0060] Hereinafter, if there is no special statement, the terms "HB3" and "thio-HB3" bo...
Embodiment 2
[0061]Example 2 Ability of HB3-1 to bind to HER2 polypeptide
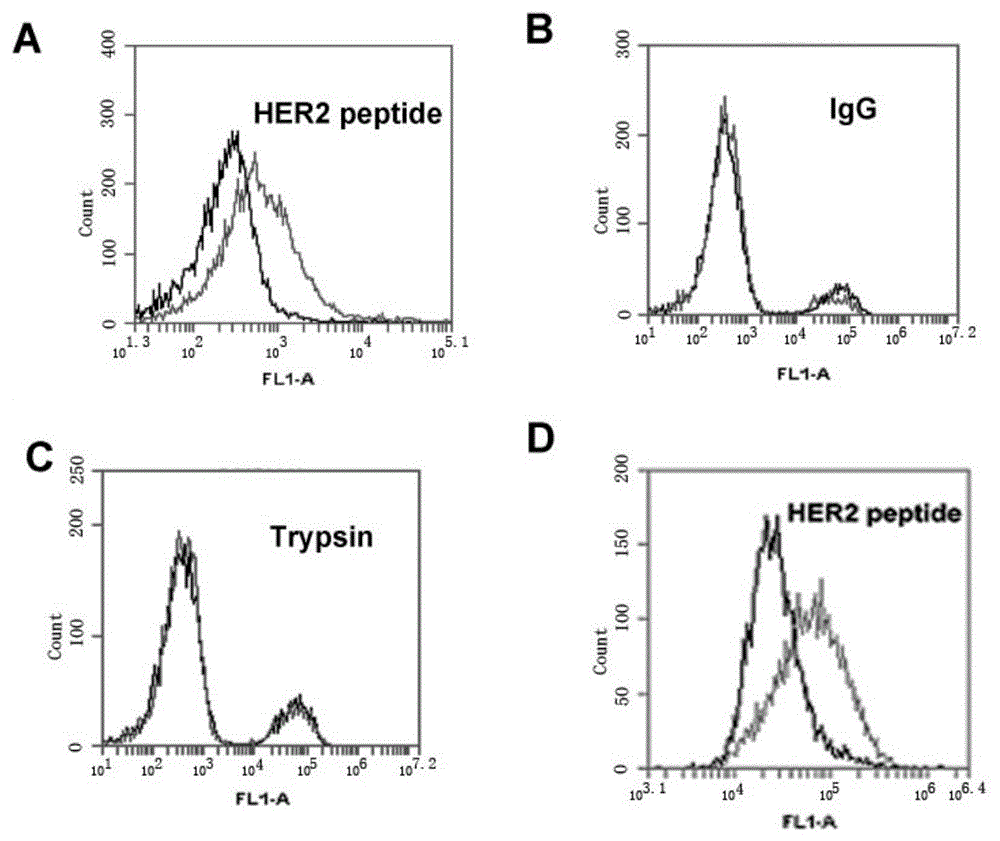
[0062] HB3-1 was modified with biotin (biotin) at the 3' end and FITC at the 5' end, and mixed with HER2 polypeptide, trypsin and The IgG-coated magnetic beads were reacted at 37°C for 30 minutes, washed twice with PBS, and analyzed by flow cytometry. The primary structure of the modified HB3-1 is as follows:
[0063] 5'FITC-GTGTGTCTGCTATTTATTTTGCTTGATTATCTCTTAGGGATTT-biotin-3'. Base A in the sequence is thio-modified.
[0064] Under the same conditions, we detected the binding of base A to the HER2 polypeptide without thio-modification.
[0065] The result is as figure 2 , wherein (A) the combination of HER2 polypeptide-coated magnetic beads and FITC-labeled HB3-1; (B) the combination of IgG antibody-coated magnetic beads and FITC-labeled HB3-1; (C) trypsin-coated Binding of magnetic beads and FITC-labeled HB3-1; (D) Binding of HER2 polypeptide-coated magnetic beads and FITC-labeled non-sulfo-modified HB3-1....
Embodiment 3
[0066] Example 3 Ability of HB3-1 to bind to HER2-positive tumor cells
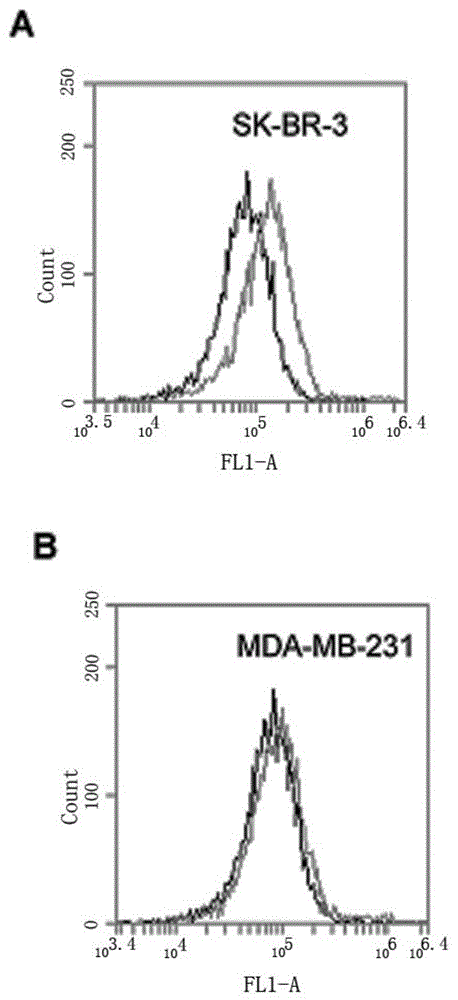
[0067] SK-BR-3 cells were cultured in modified RPMI1640 containing 15% FBS, MDA-MB-231 cells were cultured in DMEM high-glucose medium containing 10% FBS, 100 U / ml penicillin and 100 mg / ml streptomycin, All cells were stored at 37°C, 5% CO 2 Cultured in an incubator, the cells used in the following experiments were all in the logarithmic growth phase.
[0068] Scrape 5×10 separately 5 The SK-BR-3 and MDA-MB-231 cells were reacted with HB3-1 (modified biotin at the 3' end and fluorescein at the 5' end) in 200 μl binding buffer (PBS) at 37°C for 30 min, Wash twice with PBS and analyze by flow cytometry. see results image 3 , (A) is HER2 positive SK-BR-3 cells. (B) HER2-negative MDA-MB-231 cells. The black curve is the control fluorescence signal generated by random library single-stranded DNA, and the gray curve is the fluorescence signal generated by sulfo-HB3-1. The results in the accompanying dra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com