Double digital PCR fluorescent quantitative detection method for transgenic maize MON810
A technology for fluorescent quantitative detection and genetically modified corn, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve the problem that genetically modified detection is in its infancy, unsuitable for genetically modified component detection methods, and deviations in experimental results and other problems, to achieve the effect of simple and easy detection process, avoid adverse effects, and improve stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
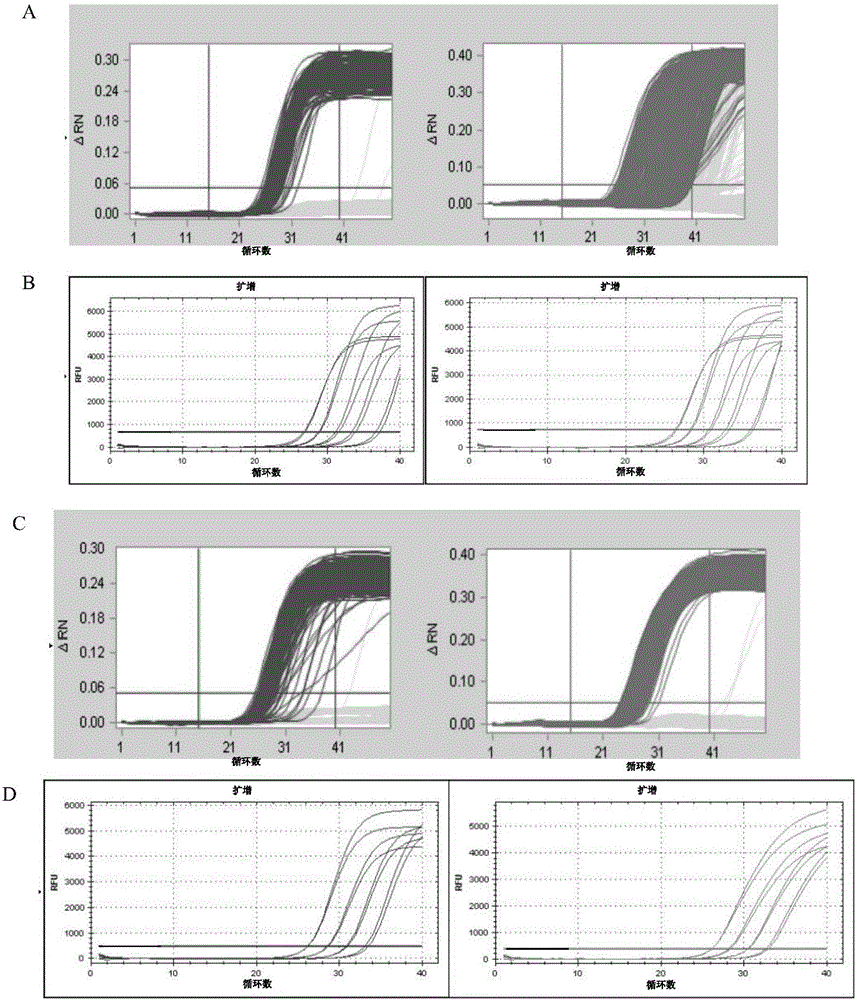
[0032] Example 1 Design of primers and probe combinations for double digital PCR fluorescence quantitative detection of transgenic maize MON810
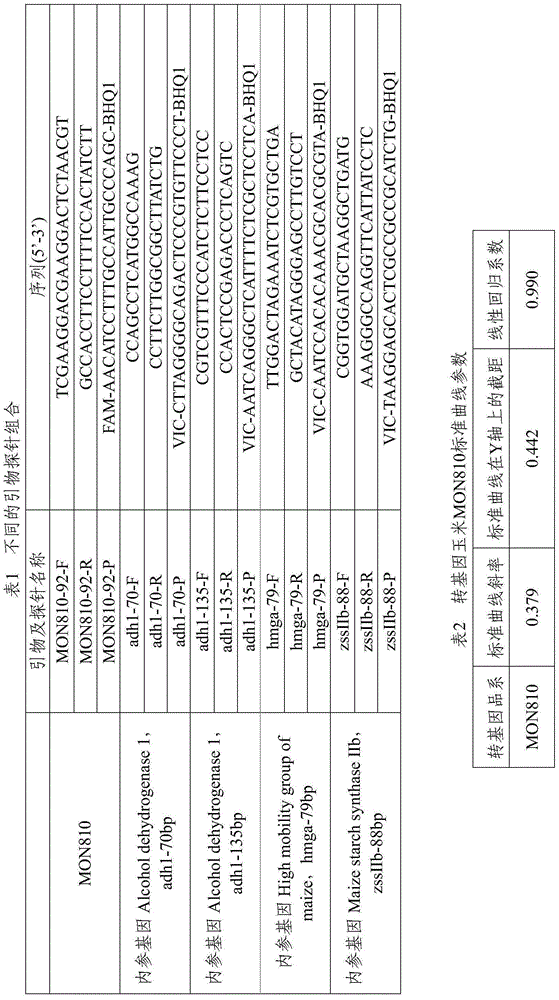
[0033] According to the insertion sequence and border sequence of the exogenous gene of the transgenic maize MON810 strain, the following primer and probe combinations (SeqIDNo.1-6) for the dual digital PCR fluorescence quantitative detection of the transgenic maize MON810 were designed:
[0034] Foreign gene forward primer: 5'-TCGAAGGACGAAGGACTCTAACGT-3'
[0035]Foreign gene reverse primer: 5'-GCCACCTTCCTTTTCCACTATCTT-3'
[0036] Foreign gene probe: 5'-FAM-AACATCCTTTGCCATTGCCCAGC-BHQ1-3'
[0037] Internal reference gene hmga forward primer: 5'-TTGGACTAGAAATCTCGTGCTGA-3'
[0038] Internal reference gene hmga reverse primer: 5'-GCTACATAGGGAGCCTTGTCCT-3'
[0039] Internal reference gene hmga probe: 5'-VIC-CAATCCACACAAACGCACGCGTA-BHQ1-3'.
Embodiment 2
[0040] Example 2 Establishment of double digital PCR quantitative detection method for transgenic maize MON810
[0041] 1.1 Experimental materials
[0042] The transgenic maize MON810 sample used in this example and its parental non-transgenic samples, transgenic maize lines NK603, MON863, Bt176, MIR162, 3272, and MIR604 were all provided by the Chinese Academy of Inspection and Quarantine. The PCR chip was purchased from Fluidigm, USA, model number 48.770 DigitalArrayChip; the BioMarkHD high-throughput gene analysis system (BiomarkHD System) was purchased from Fluidigm.
[0043] 1.2 Genomic DNA extraction of transgenic crop seed samples for experiments
[0044] (1) The crop seed sample is ground and thoroughly air-dried;
[0045] (2) Mixing the transgenic corn line and parental non-transgenic seed samples in different proportions, and mixing overnight with a mixing instrument to obtain samples with different transgene concentrations for experiments;
[0046] (3) Weigh 100 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com