A kind of cotton ATP hydrolase atpase 2 and its coding gene and application
A technology of hydrolase and gene, applied in the field of cotton ATP hydrolase ATPase‑2 and its coding gene and application, capable of solving problems such as complex mechanism and important issues of plant salt resistance to be explored
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1. Cotton SSH library construction under salt stress:
[0019] The specific method is:
[0020] According to Clontech's PCR-select TM The method shown in the cDNA Subtraction Kit kit manual was used to construct the SSH library (suppression subtraction library) by the suppression subtractive hybridization method. During the experiment, the mRNA extracted from the salt-treated cotton root tissue was used as the sample (Tester), and the mRNA extracted from the untreated cotton root tissue was used as the control (Driver). Specific steps are as follows:
[0021] (1) Test materials:
[0022] African cotton (National Cotton Medium-Term Bank, acquired by China Cotton Research Institute, unified number: ZM-06838) was sown on sterilized vermiculite, cultivated at 25°C, with a light-dark cycle of 16h / 8h, and watered 1 / 2 2MS medium (9.39mM KNO 3 , 0.625mM KH2PO 4 , 10.3mM NH 4 NO 3 , 0.75mM MgSO 4 , 1.5mM CaCl 2 , 50 μM KI, 100 μM H 3 BO 3 , 100 μM nSO 4 , 3...
Embodiment 2
[0033] Example 2 Cloning of cotton ATP hydrolase gene GhATPase-2
[0034] After removing redundant DNA from the clone of the identified cotton SSH library from colony Gh-S2-176, the sequence is SEQ ID No: 3, and sequence analysis shows that the protein encoded by this sequence belongs to ATP hydrolase. In this paper, the full-length coding gene corresponding to the sequence of SEQ ID No: 3 is named GhATPase-2, and the corresponding protein is named ATPase-2.
[0035] SEQ ID No: 3:
[0036]
[0037]
[0038] Cloning of the full-length coding gene of GhATPase-2
[0039] According to the obtained sequence of SEQ ID No: 3, the following two specific primers were designed as the 5' end specific primers of 3' RACE.
[0040] GhATPase-2 GSP1: SEQ ID No: 4:
[0041] GTAGAAGCTT GCATAAATGC TG
[0042] GhATPase-2 GSP2: SEQ ID No: 5:
[0043] ACAGCAAAGG CTATAGCAAC TGA
[0044] The experimental steps were operated according to the instructions of the kit (the 3'RACE System for R...
Embodiment 3
[0079] The construction of the plant expression vector of embodiment 3 GhATPase-2 gene
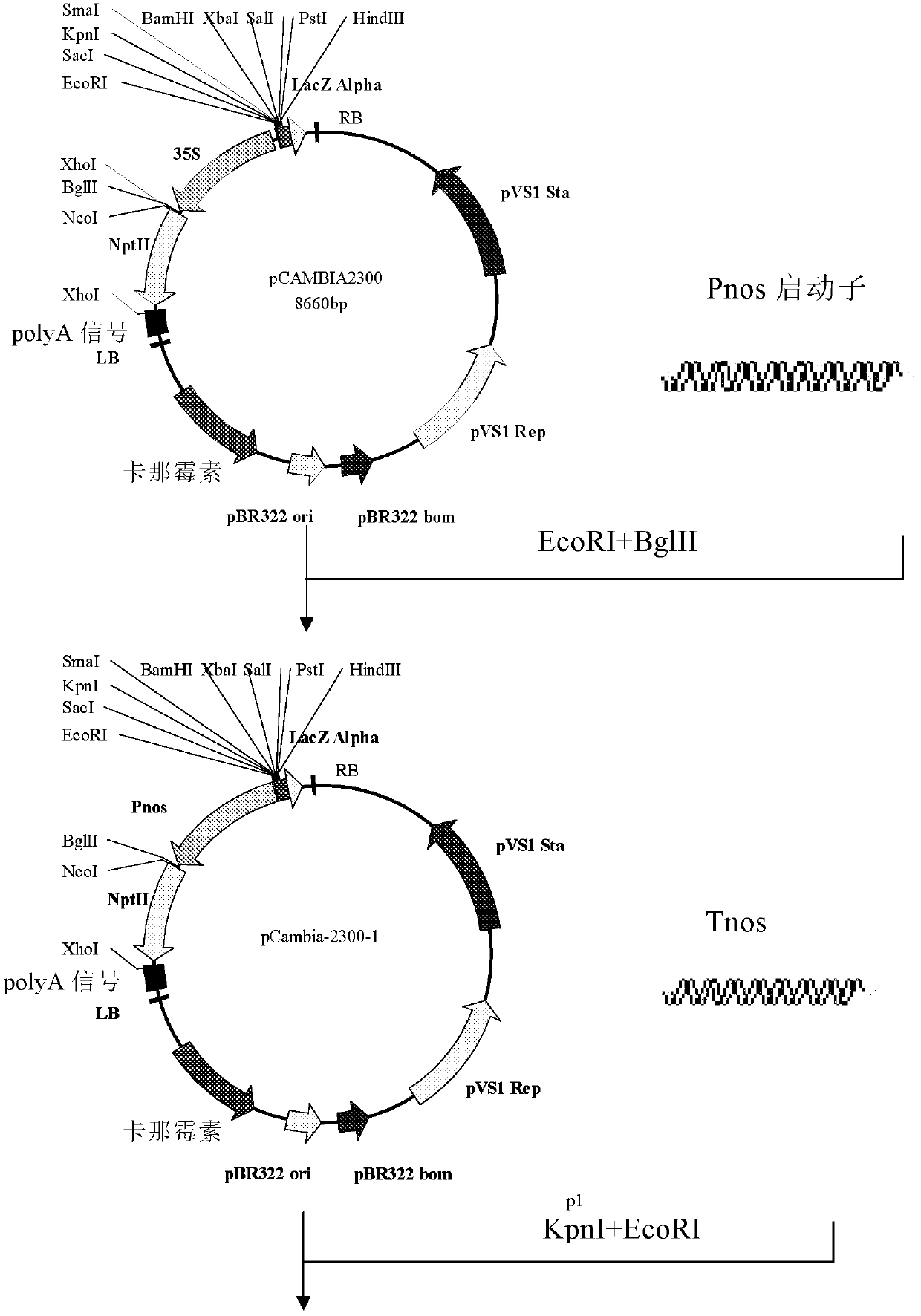
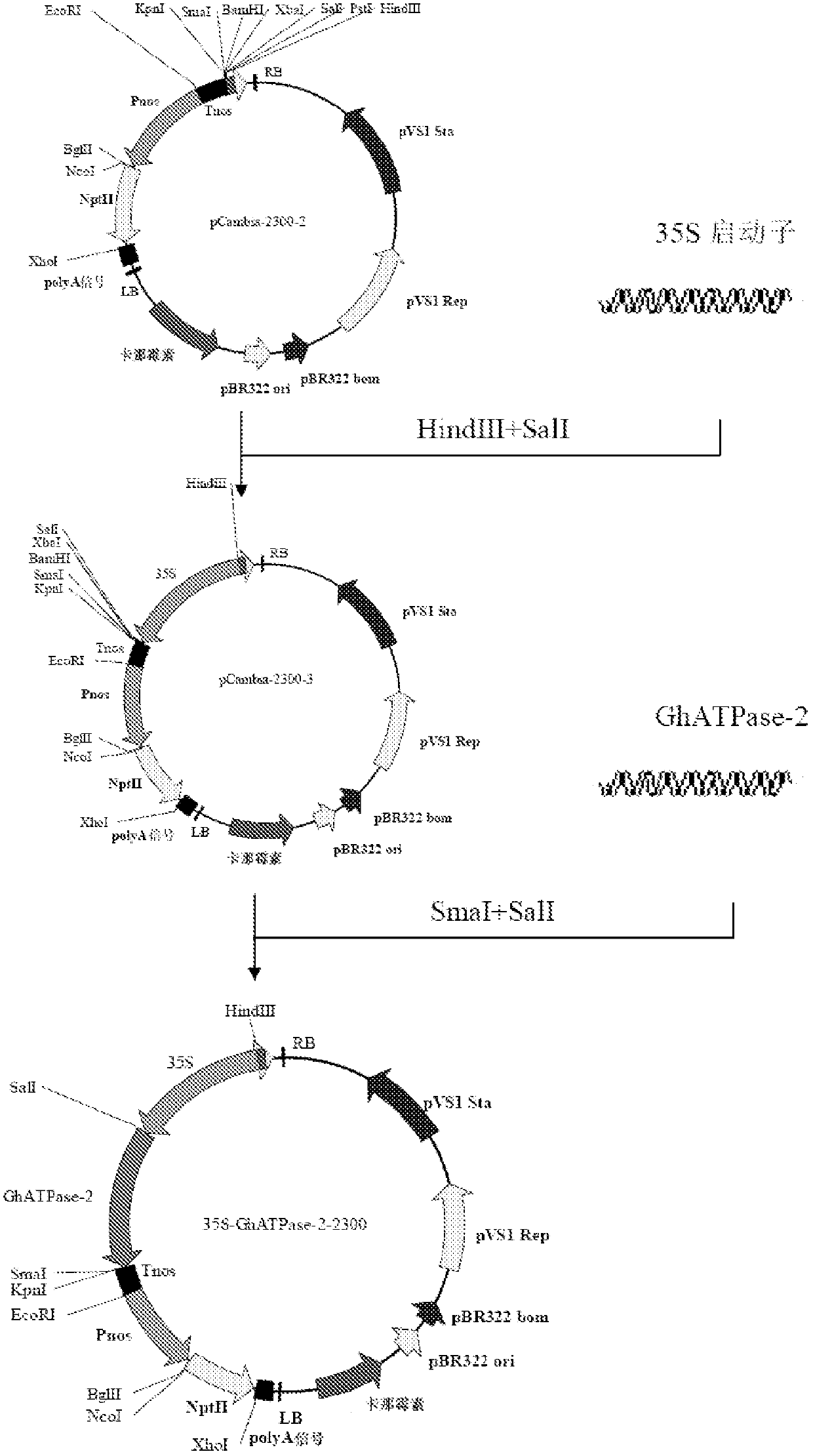
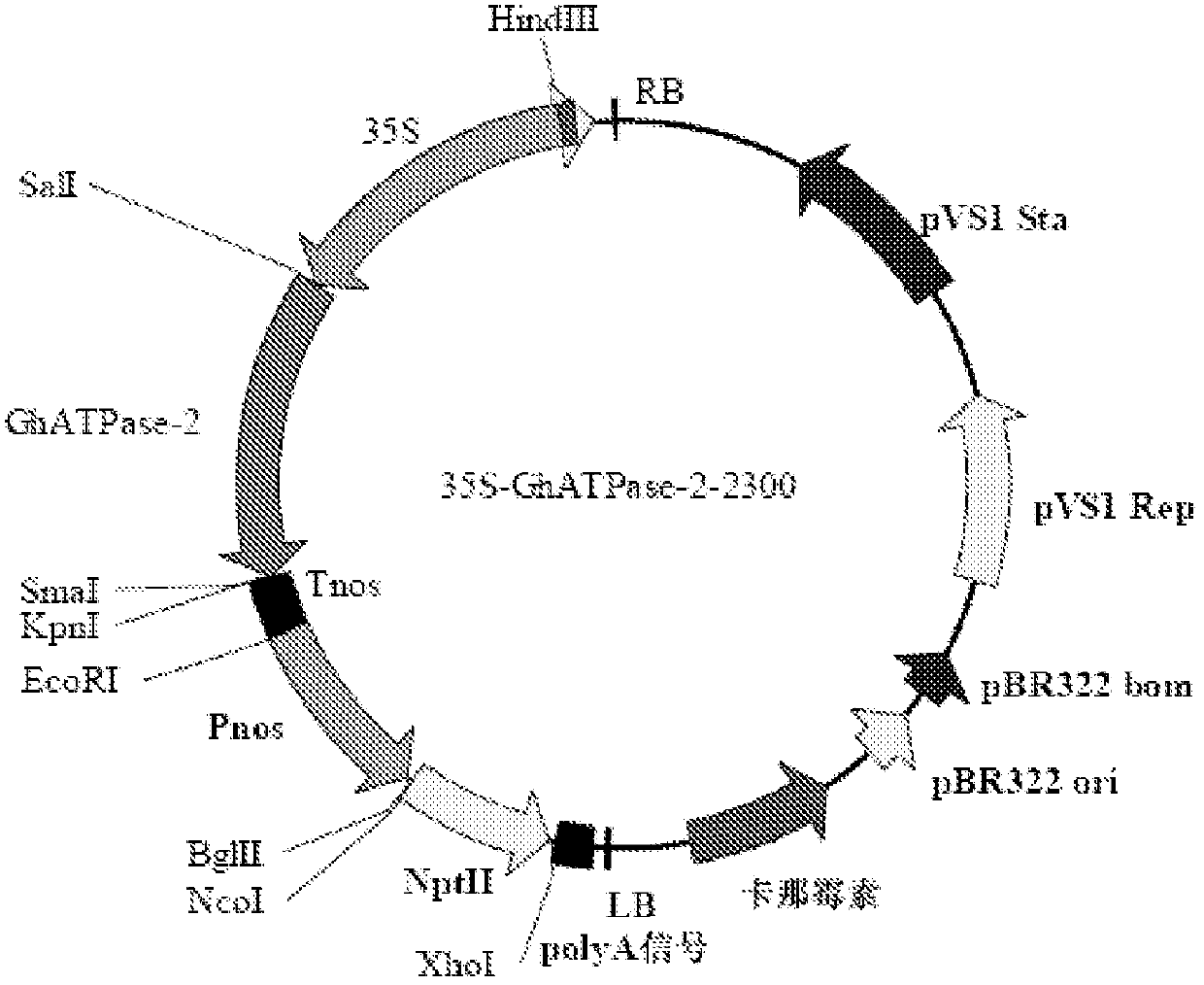
[0080] The plant binary expression vector pCAMBIA2300 (purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd.) was selected as the plant expression vector, and the 35S promoter of the NPTII gene containing double enhancers was replaced with the Pnos promoter to reduce the expression of NPTII protein in plants . The 35S promoter and the Tnos terminator were selected as the promoter and terminator of the GhATPase-2 gene, respectively, and the construction flow chart is shown in Figure 1.
[0081] Primers SEQ ID NO: 12 and SEQ ID NO: 13 were used to amplify Pnos with the plant expression vector pBI121 plasmid (purchased from Beijing Huaxia Ocean Technology Co., Ltd.) as a template, and TaKaRa's PrimeSTAR HS DNA polymerase was used. 50 μl PCR reaction system: 10 μl 5×PS Buffer, 3 μl 2.5 mM dNTP, 1.0 μl pBI121 plasmid, 1.0 μl PrimeSTAR HS DNA polymerase, 10 μM primers SEQ ID NO: 12 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com