SNP (single nucleotide polymorphism) marker for distinguishing Chinese population and Japan population of haliotis discus hannai
A wrinkled abalone and colony technology, applied in recombinant DNA technology, biochemical equipment and methods, DNA/RNA fragments, etc., to achieve the effects of simple operation, low cost and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
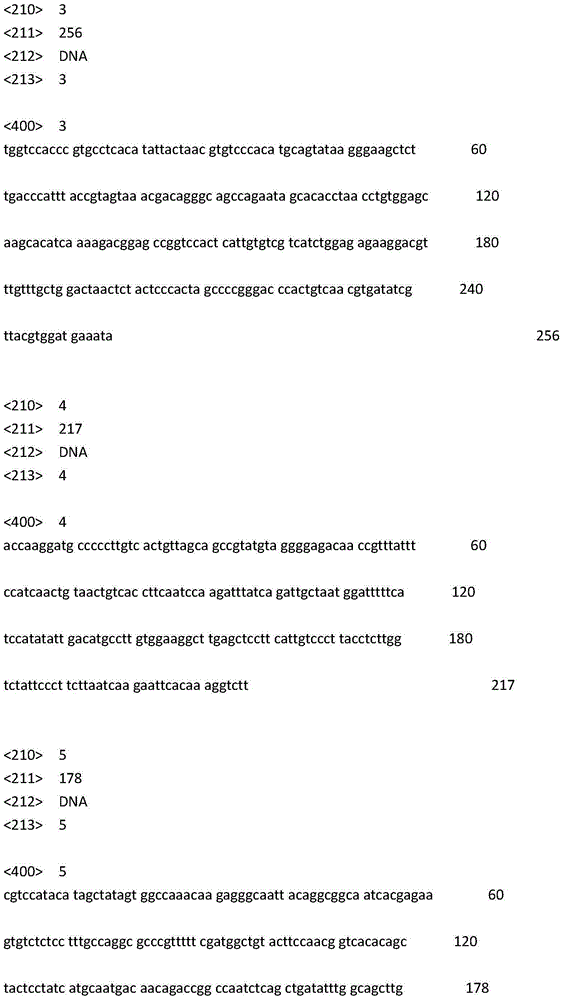
[0025] Using next-generation sequencing technology, high-throughput sequencing was performed on 5 Chinese individuals and 5 Japanese individuals of Abalone rugosa, and the sequencing reads of all individuals were used to construct the genome scaffold of Abalone rugosa, which was used as a reference genome sequence for subsequent analysis. Compare the sequencing reads of Chinese individuals and Japanese individuals to the reference genome, screen the SNP sites and calculate their allele frequencies in the Chinese and Japanese populations, and use the Chi-squared test to calculate the alleles of the two populations The difference in frequency is used to screen the SNP sites with significant differences. A total of 5 SNP sites were screened out, wherein the site HdhSNP30_CT is the 99th position of SEQIDNO:1, and its base is C / T; the site HdhSNP32_CT is the 138th position of SEQIDNO:2, and its base is C / T; The site HdhSNP36_CA is the 107th position of SEQIDNO:3, and its base is C / ...
Embodiment 2
[0033] This embodiment uses the primer set and probe pair of the above-mentioned SNP site to verify the identification of Chinese and Japanese populations of Abalone wrinkled, including the following steps:
[0034] 1) Genomic DNA extraction of wrinkled abalone: Take about 0.1g of foot muscle, add 500μl STE lysis buffer, cut into pieces, add 50μl 10% SDS, 5μl proteinase K (20mg / ml) in turn, lyse at 56°C for about 3 hours, until the lysate is clear . Add the same volume of Tris saturated phenol (250 μl), chloroform / isoamyl alcohol (24:1) (250 μl), shake gently for 20 minutes, and centrifuge at 12000 rpm for 10 minutes. Take the supernatant and repeat the above steps until there is no protein layer between the aqueous phase and the organic phase. Take the supernatant, add the same volume of chloroform / isoamyl alcohol (24:1), shake gently for 20min, and centrifuge at 12000rpm for 10min. Take the supernatant, add 1 / 10 volume of 3M NaAc (pH5.2) and 2 volumes of cold absolute et...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com