Detection kit for Neosporiasis
A technology for neosporosis and a kit, which is applied in the field of molecular biology detection, can solve the problems of unsuitable detection, cumbersome operation, and lengthy process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0026] Example 1. Design of neosporosis detection primers and fluorescent probes
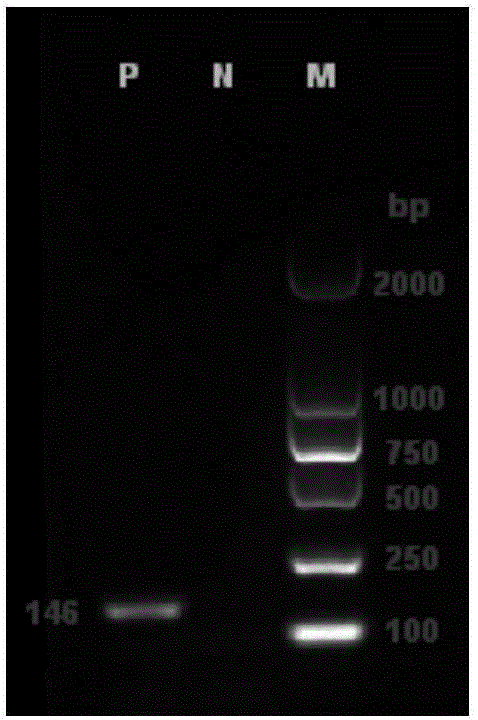
[0027] ① Refer to the literature to understand the relevant characteristics of the Neospora NcSRS2 gene, download the Neospora NcSRS2 gene sequences uploaded in GenBank from different countries and regions, use DNAMAN software to compare and analyze all the sequences, and find out the conserved sequence of the NcSRS2 gene. The detection primers and fluorescent probes were designed within the sequence, and the design principles followed the Real-TimePCR primer amplification product length of 80-150bp. ② Use PrimerExpress3.0 and Oligo6.24 software to design detection primers NcSRS2F / R and MGB fluorescent probe NcSRS2P according to the sequence of the conserved region of Neospora NcSRS2 gene (accession number: U93870.1), and the 5' end of the probe is labeled with FAM fluorescent reporter Group, 3' labeled MGB fluorescence quencher group, amplified 146bp target fragment. The upstream primer NcSRS2...
example 2
[0028] Example 2: Preparation of target gene fragment positive plasmid
[0029] ① First, the designed detection primers were entrusted to Beijing Dingguo Changsheng Biotechnology Co., Ltd. for synthesis, and the synthesized primers were used for routine PCR amplification of Neospora positive DNA. The conventional PCR reaction system was 25 μL: 10×PCRBuffer (Mg 2+ plus) 2.5 μL, dNTP (2.5mM) 2 μL, upstream and downstream primers (10pM) 1 μL each, TaqDNA polymerase (5U / μL) 0.15 μL, template DNA 1.5 μL, ddH 2 O to make up 25 μL. The cycle conditions were: pre-denaturation at 94°C for 5min; denaturation at 94°C for 30s, annealing at 56°C for 30s, extension at 72°C for 30s, 35 cycles; extension at 72°C for 5min. ② Take 1 μL of 6×DNALoadingBuffer and add it to 5 μL of PCR product and mix well, and analyze the size of the amplified band by electrophoresis on 1.5% agarose gel and 100V for 40 minutes. If the band size is 146bp, follow-up experiments can be continued (see attached fig...
example 3 3D
[0030] Example 3. 3D digital PCR screening for optimal annealing-extension temperature and absolute quantitative analysis of amplified templates
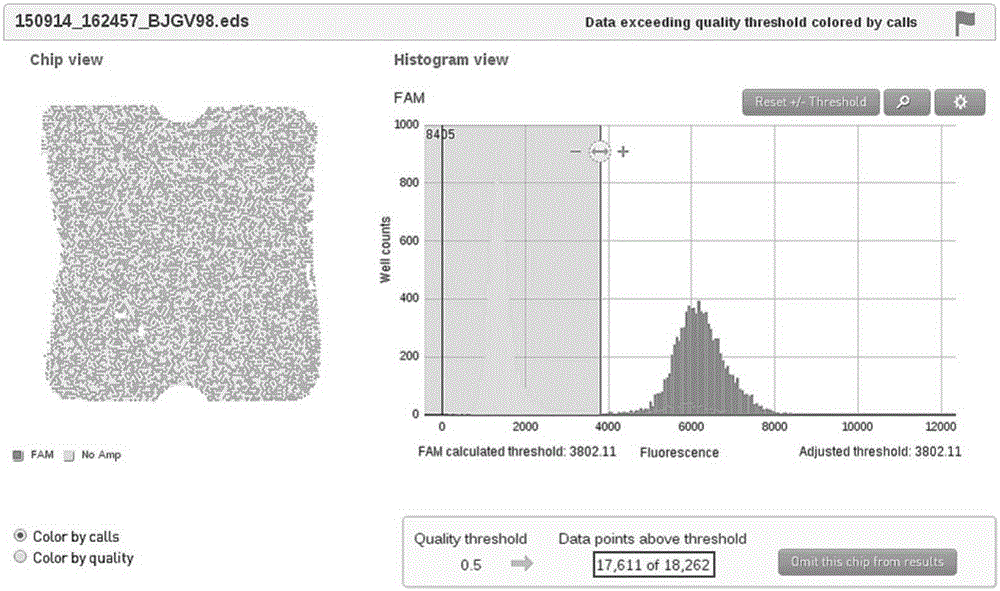
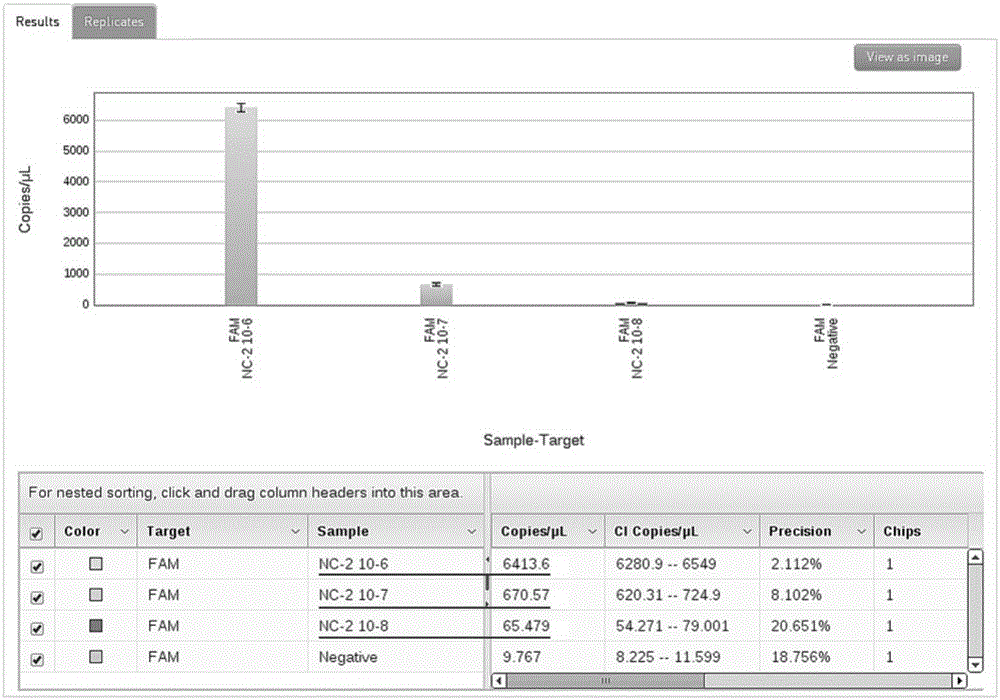
[0031] ① Using NcSRS2F / R and NcSRS2P to extract 10 positive plasmids (32.5ng / μL) -6 、10 -7 、10 -8 Dilute the titer for 3D digital PCR amplification to screen the optimal annealing-extension temperature of primers and probes and determine the concentration of templates involved in the reaction. ② 3D digital PCR reaction system is 15 μL: 2×3DMasterMix 7.5 μL, primers, probes (10pM) 1 μL each, plasmid template 2 μL, ddH 2 O to make up 15 μL. The optimal cycle conditions for screening were: pre-denaturation at 96°C for 10 minutes; annealing-extension at 58°C for 2 minutes, denaturation at 98°C for 30 seconds, 40 cycles; extension at 60°C for 2 minutes. Therefore, 58°C is used as the optimal annealing-extension temperature for Real-TimePCR amplification (see attached figure 2 ); yields 10 -6 In the diluted plasmid template, the te...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com