Method for high flux synthesis of nucleotide pool and assembling of double-stranded DNA from semiconductor chips
A nucleotide pool and semiconductor technology, applied in the field of molecular biology, can solve the problems of high cost, cumbersome steps, low degree of integration and miniaturization, etc., and achieve the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
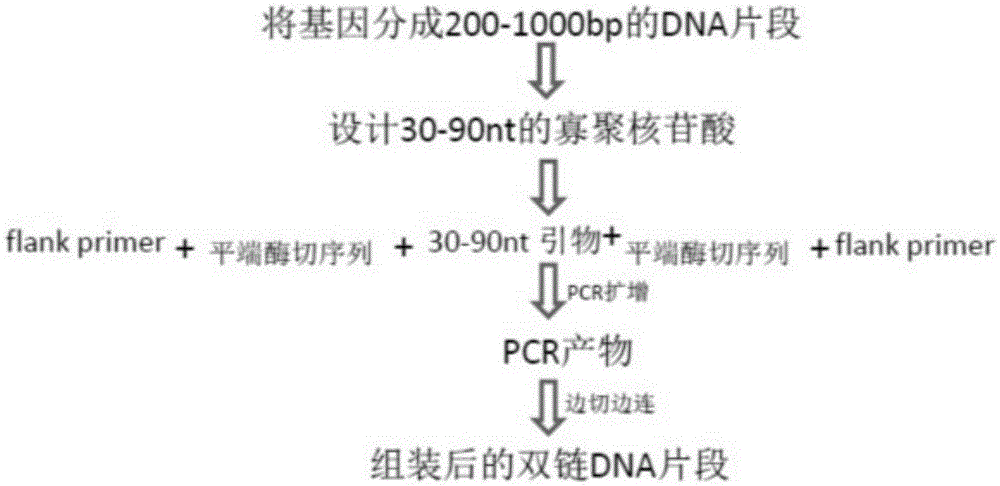
[0033] Such as figure 1 As shown, a method for high-throughput synthesis of a semiconductor chip nucleotide pool and assembled double-stranded DNA is disclosed in Embodiment 1, the method comprising the following steps:
[0034] (1) Divide the pre-synthesized double-stranded DNA into multiple 200-1000bp DNA fragments.
[0035] (2) Design nucleotides with the DNA fragment described in (1), the length of each nucleotide is 30-90nt, the last 4 bases and the next nucleotide at the 3' end of each nucleotide The first 4 nucleotide sequences of the 5' end are the same.
[0036] (3) Add a 20nt flank sequence to the 30-90nt nucleotide ends designed in (2), and the restriction endonuclease MlyI restriction endonuclease recognition sequence of Type II, and synthesize a total length of 80-200nt Nucleotide sequence, the nucleotide sequence is prepared with a semiconductor chip; In the present embodiment, the TypeII restriction endonuclease can be all TypeII restriction endonucleases that...
Embodiment 2
[0041] In Example 2, using the method disclosed in Example 1, a semiconductor chip was used to synthesize a nucleotide pool and use it as a raw material to assemble double-stranded DNA.
[0042] 步骤1:将预合成的双链DNA分成多个DNA片段,DNA片段的序列如下:GGATCCAGAGCAGAGGAGCCAGCTGTGGGCACCAGTGGCCTCATCTTCCGAGAAGACTTGGACTGGCCTCCAGGCAGCCCACAAGAGCCTCTGTGCCTGGTGGCACTGGGCGGGGACAGCAATGGCAGCAGCTCCCCCCTGCGGGTGGTGGGGGCTCTAAGCGCCTATGAGCAGGCCTTCCTGGGGGCCGTGCAGAGGGCCCGCTGGGGCCCCCGAGACCTGGCCACCTTCGGGGTCTGCAACACCGGTGACAGGCAGGCTGCCTTGCCCTCTCTACGGCGGCTGGGGGCCTGGCTGCGGGACCCTGGGGGGCAGCGCCTGGTGGTCCTACACCTGGAGGAAGTGACCTGGGAGCCAACACCCTCGCTGAGGTTCCAGGAGCCCCCGCCTGGAGGAGCTGGCCCCCCACTCGAG
[0043] Step 2: Design primers, the designed primers are as shown in Table 1:
[0044] Table 1 Primer list designed
[0045]
[0046] Step 3: Add a 20nt flank sequence and a restriction enzyme recognition site sequence to both ends of the primer designed in step 2. In this example, the restriction endonuclease MlyI is used as an example, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com