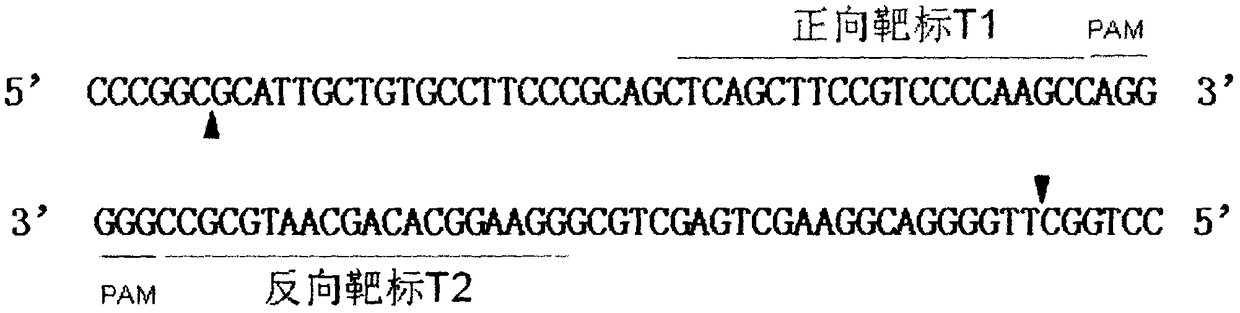A pair of sgRNAs targeting the porcine rela gene
One gene, one pair technology, applied in the field of sgRNA targeting recognition of porcine RelA gene
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1, the construction of sgRNA expression plasmid pair
[0021] 1. sgRNA target design
[0022] The coding region (959-1033, as shown below) of the tenth exon of the porcine RelA gene was extracted from the sequence with the NCBI accession number NM_001114281.1, using the MIT online software (http: / / crispr.mit.edu / ) Design the target site.
[0023] GACCCACCGACCCCCC GGCCTGCAAC CCGGCGCATT GCTGTGCCTT CCCGCAGCTCAGCTTCCGTC CCCAAGCCAG
[0024] The forward target sequence is TCAGCTTCCGTCCCCAAGCC (54-73) and the reverse target sequence is GGAAGGCACAGCAATGCGCC (28-47). The two target sequences are arranged in a "head-to-head" manner, and the distance between the two is 6 bp, that is, there is a 6 bp interval. Name them as forward target T1 and reverse target T2 respectively. target design as figure 1 shown.
[0025] 2. Construction of sgRNA expression plasmid pair
[0026] First, according to the designed target sequence, send it to Beijing Liuhe Huada Gene Tech...
Embodiment 2
[0032] Example 2. Verifying the biological activity of the sgRNA expression plasmid pair constructed in Example 1 by sequencing
[0033] PEF cells (porcine fetal fibroblasts): PEF cells were isolated from aborted pig fetuses (see references for the isolation method: Li Hong, Wei Hongjiang, Xu Chengsheng, Wang Xia, Qing Yubo, Zeng Yangzhi; Banna Miniature Pig Inbred Line The establishment of fetal fibroblast cell line and its biological characteristics; Journal of Hunan Agricultural University (Natural Science Edition); Vol. 36 No. 6; December 2010; 678-682).
[0034] 1. Co-transfect 4 μg of each of the recombinant plasmids pX335-sgRNA-F and pX335-sgRNA-R into 1×10 6PEF cells to obtain recombinant cells. The specific steps of transfection are as follows: use a nuclear transfer instrument (Amaxa, model: AAD-1001S) and a matching mammalian fibroblast transfection kit (Amaxa, product number: VPI-1002) for transfection. First, use 0.05% trypsin (Gibco, catalog number: 610-5300AG)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com