PCR-HRM primer for quickly detecting salmonella gallinarum and application thereof
A PCR-HRM and Salmonella technology is applied in the field of PCR-HRM primers for rapid detection of Salmonella typhimurium.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
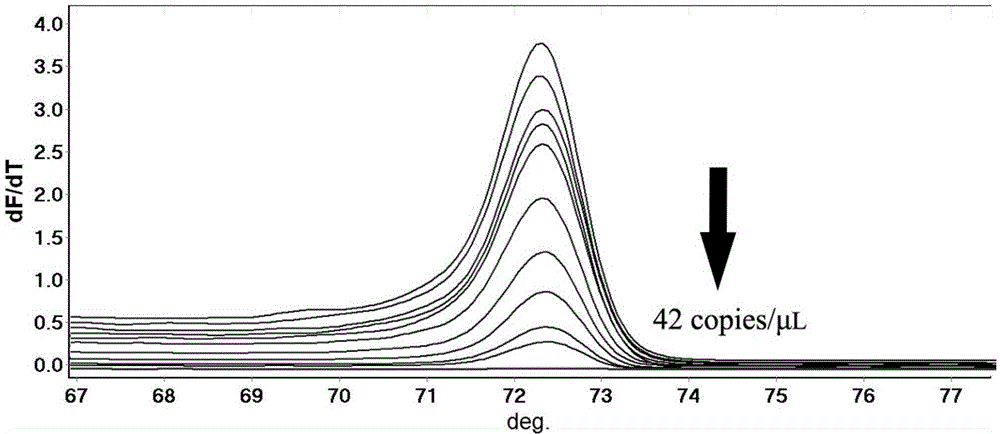
[0035] Example 1 is used to detect Salmonella gallinarum typhi HRM primers and reaction conditions
[0036] (1) Primers: According to the complete genome sequence of Salmonella published by NCBI and related information, it is found that Salmonella gallinarum typhi has a regular change at position 598 on the rfbS gene. G, some other serotypes and strains of other types do not contain this gene or have large changes, so specific amplification primers can be designed according to this site. Adopt Oligo7 primer design software, according to the Salmonella rfbS sequence issued by GeneBank (the GeneBank accession number of the rfbS sequence of Salmonella pullorum: LK931482; the GeneBank accession number of the rfbS sequence of Salmonella gallinarum typhi: AF442573; ) and the rfbS sequence obtained by sequencing the Salmonella typhi standard strain (the rfbS sequence is the same as that in GeneBank accession number: AF442573), and a primer pair was designed on both sides of the 598 s...
Embodiment 2
[0042] Embodiment 2 is used to detect the application method of the PCR-HRM primer of Salmonella gallinarum typhi
[0043] (1) Preparation and PCR-HRM analysis of Salmonella typhi standard substance
[0044] (1) Extraction of bacterial DNA:
[0045] Resuscitate the standard strain of Salmonella typhi (CICC21510), pick a single colony and shake the bacteria in 1mL LB medium for 12 hours, use the OmegaDNAkit kit to extract the bacterial DNA, follow the instructions to extract and purify the sample DNA, and store the extracted DNA at -20°C .
[0046] (2) Preparation of standard products:
[0047] In order to verify the feasibility of the primers and methods of the present invention, construct a positive sample at the same time, provide a positive template for subsequent clinical sample detection, use the extracted standard strain DNA as a template, use the upstream primer Sg-up described in Example 1, the downstream Primer Sg-low was used for ordinary PCR amplification.
[00...
Embodiment 3
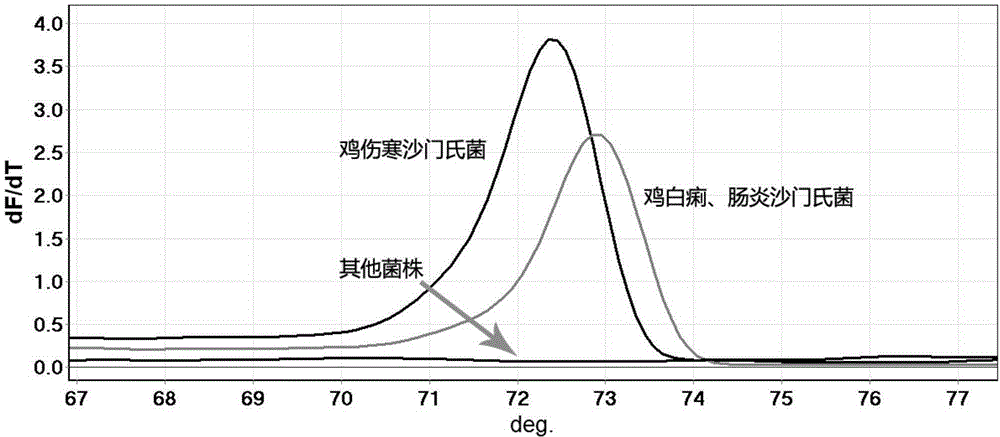
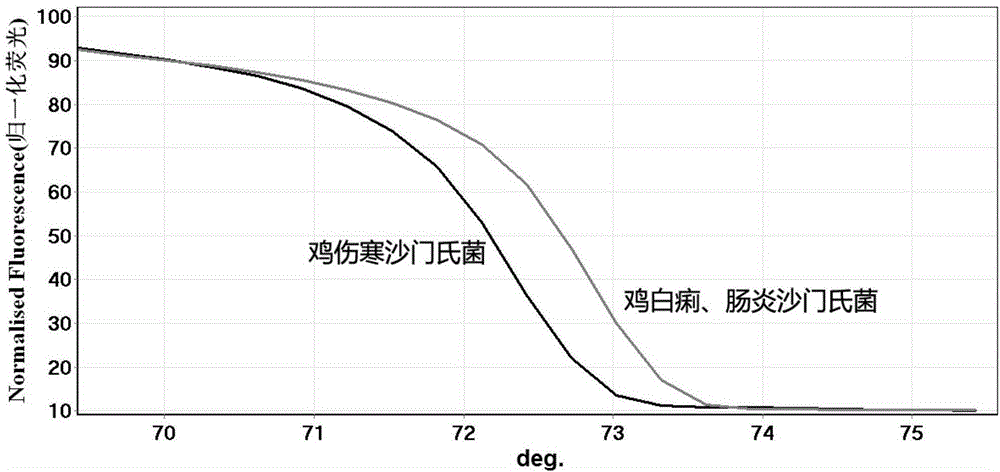
[0066] Example 3 is used to detect the clinical application of Salmonella gallinarum typhi HRM primer
[0067] Liver samples were collected from clinically suspected Salmonella typhi-affected chickens, and a total of 15 samples were collected.
[0068] (1) Processing of clinical samples
[0069] Take 25g of the liver of the tested chicken, grind the sample into a sterile enrichment bag, add 225g of BPW (protein water) to pre-enrichment in a constant temperature shaker at 37°C at 100r / min for 6-8 hours, take 1mL of the pre-enrichment solution, Add 9 mL of selective enrichment solution TTB (sodium tetrathiosulfonate brilliant green), and incubate at a constant temperature of 37°C for 20 hours.
[0070] (2) Extraction of sample DNA
[0071] Take 1 mL of the selective pre-enrichment solution, and follow the instructions of the OmegaDNAkit kit to extract and purify the sample DNA, and store the extracted DNA at -20°C.
[0072] (3) PCR-HRM amplification
[0073] Using DNA as a t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com