A kind of CRISPR-Cas9 system targeting apocIII and its application
A targeted and useful technology, applied in the field of genetic engineering, can solve problems such as easy generation of drug resistance, and achieve the effect of lowering blood lipid levels and reducing recovery.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Design and synthesis of sgRNA specifically targeting apoCIII gene in CRISPR-Cas9 specific knockout of human apoCIII gene
[0032] 1. Design of sgRNA targeting human apoCIII gene
[0033] (1) Select the sequence of 5'-GGN(19)GG or 5'-GN(20)GG or 5'-N(21)GG on the apoCIII gene.
[0034] (2) The target site of sgRNA on the apoCIII gene is located in the exon of the gene.
[0035] (3) The targeting site of sgRNA on the apoCIII gene is located on the common exons of different splicing forms.
[0036] (4) Use BLAT in the UCSC database or BLAST in the NCBI database to determine whether the target sequence of the sgRNA is unique and reduce potential off-target sites.
[0037] According to the above method, we designed a total of 37 sgRNAs targeting the human apoCIII gene, the sequences of which are shown in the sequence table SEQ ID NO.4-40.
[0038] 2. Selection of sgRNA targeting human apoCIII gene
[0039] (1) The target sequence of the sgRNA targeting the apoCIII gene s...
Embodiment 2
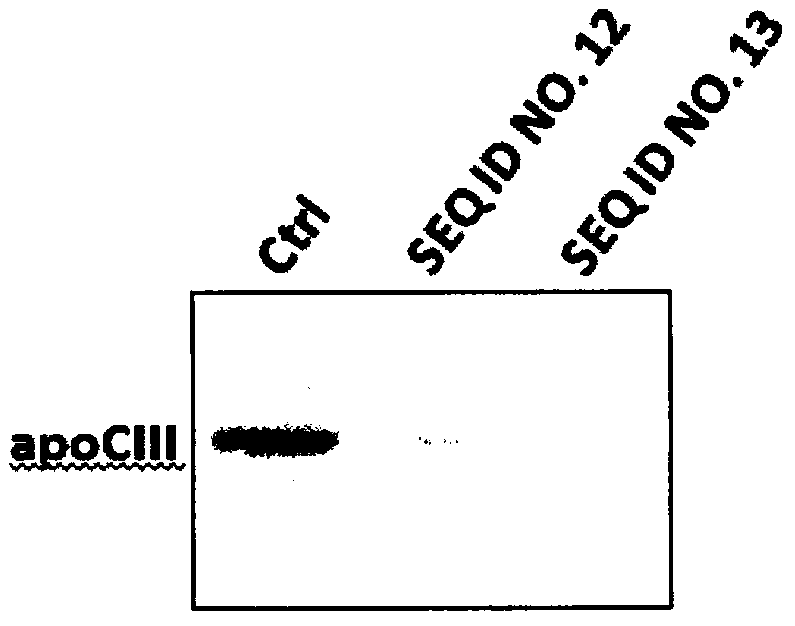
[0051] Using CRISPR-Cas9 to specifically knock out the human apoCIII gene (the sgRNA sequence used to target the apoCIII gene is shown in the sequence table as SEQ ID NO.12)
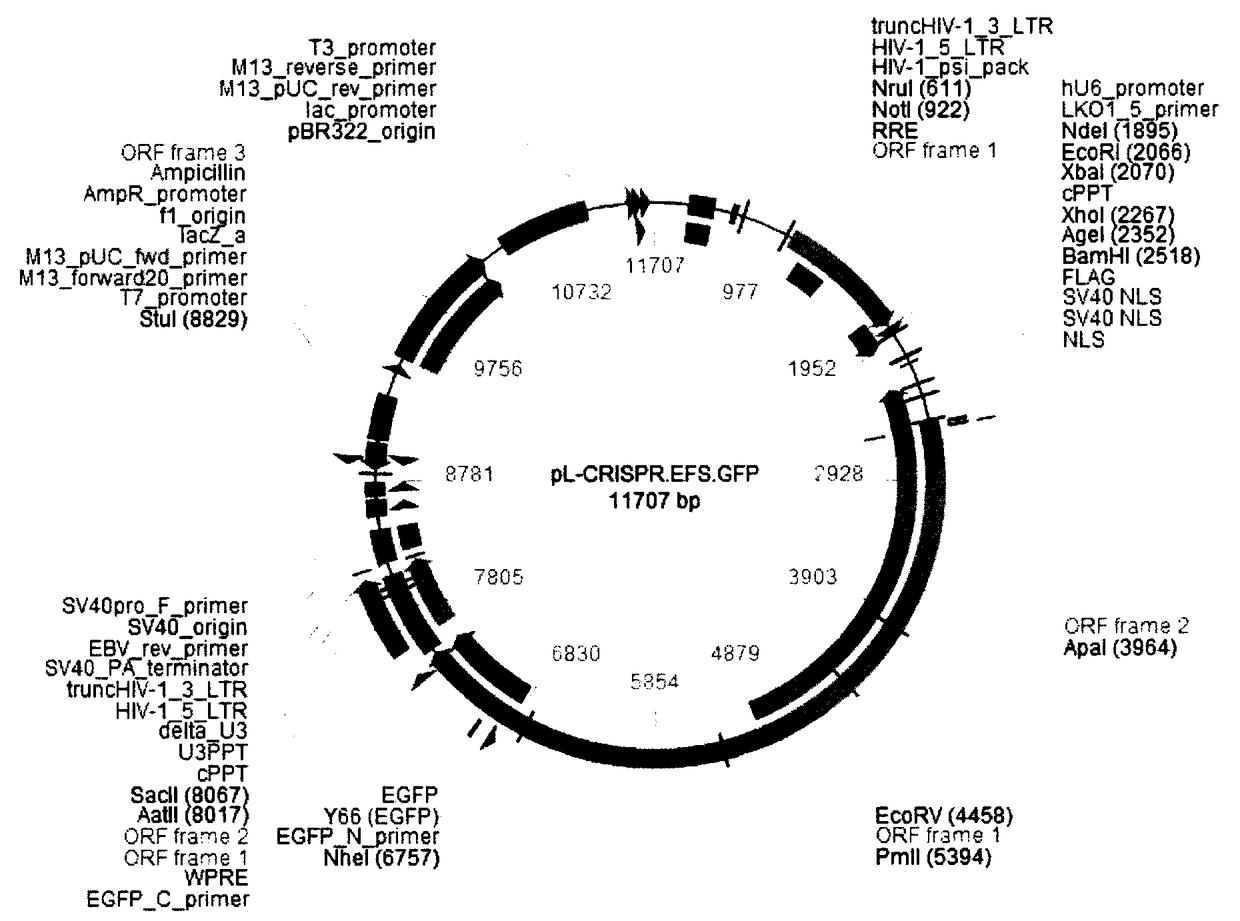
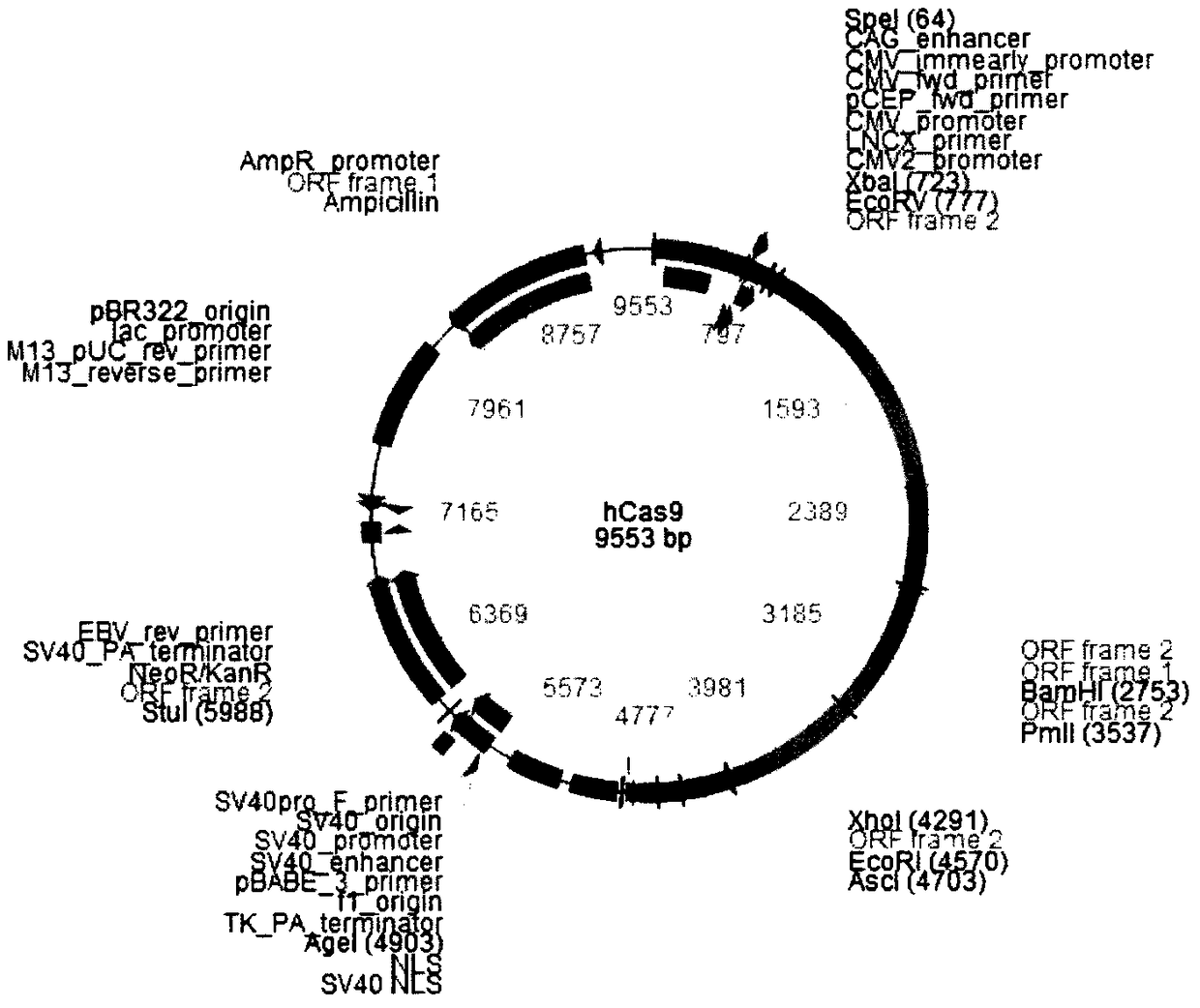
[0052] 1. Enzyme digestion of the linearized plasmid pL-CRISPR.EFS.GFP. After digestion, use AxyPrep PCR Clean up Kit (AP-PCR-250) to purify and recover to 20-40 μl sterile water.
[0053] 2. Connect the double-stranded sgRNA oligonucleotide obtained after denaturation and annealing to the linearized pL-CRISPR.EFS.GFP to obtain the pL-CRISPR.EFS.GFP-sgRNA plasmid.
[0054] 3. Transform the ligation product obtained in the above steps into competent cells and coat with Amp + Plate (50 μg / μl), and pick clones.
[0055] 4. Sequencing with universal primers to identify positive clones.
[0056] 5. Shake the bacteria at 37°C overnight to culture the positive clones, extract the plasmid, and obtain the pL-CRISPR.EFS.GFP-sgRNA plasmid.
[0057] 6. Cell culture and transfection.
[0058] 7. Neomycin screenin...
Embodiment 3
[0060] Using CRISPR-Cas9 to specifically knock out the human apoCIII gene (the sgRNA sequence used to target the apoCIII gene is shown in the sequence table as SEQ ID NO.13)
[0061] 1. Enzyme digestion of the linearized plasmid pL-CRISPR.EFS.GFP. After digestion, use AxyPrep PCR Clean up Kit (AP-PCR-250) to purify and recover to 20-40 μl sterile water.
[0062] 2. Connect the double-stranded sgRNA oligonucleotide obtained after denaturation and annealing to the linearized pL-CRISPR.EFS.GFP to obtain the pL-CRISPR.EFS.GFP-sgRNA plasmid.
[0063] 3. Transform the ligation product obtained in the above steps into competent cells and coat with Amp + Plate (50 μg / μl), and pick clones.
[0064] 4. Positive clones were identified by sequencing with universal primers.
[0065] 5. Cultivate positive clones overnight on a shaker at 37°C, and extract the plasmid with a kit to obtain the pL-CRISPR.EFS.GFP-sgRNA plasmid.
[0066] 6. Cell culture and transfection.
[0067] 7. Neomycin ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com