Microbial markers and uses therefor
A mammalian, bacterial technology, applied in the field of microbial markers and their uses, which can solve problems such as lack of detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
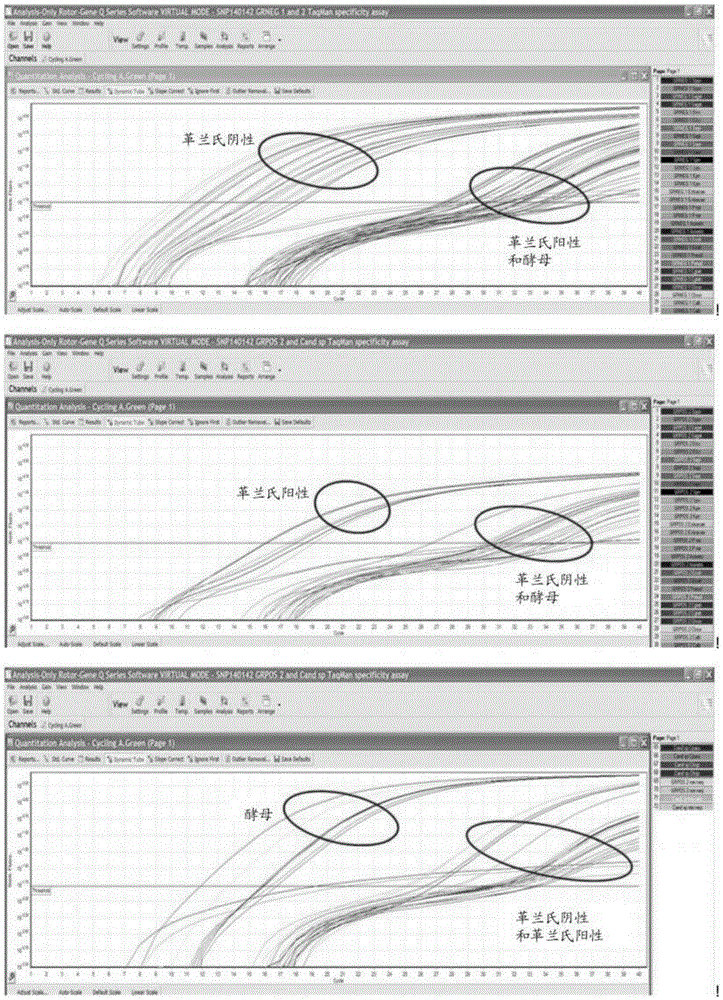
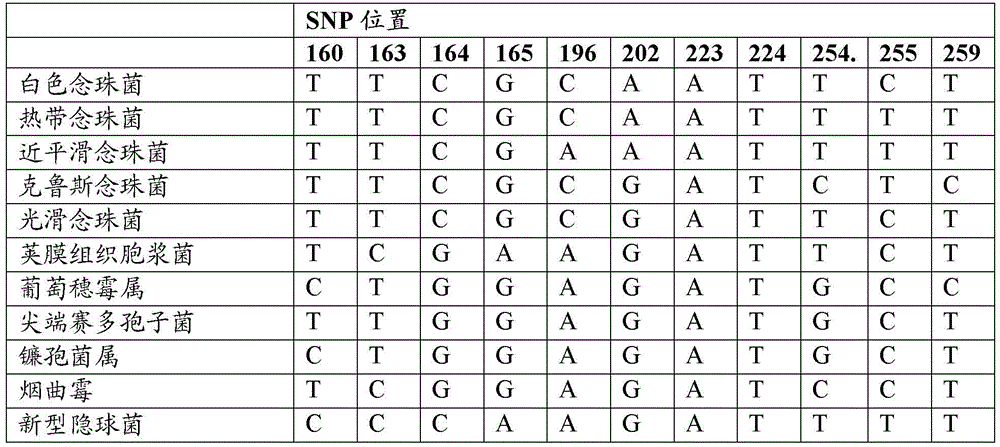
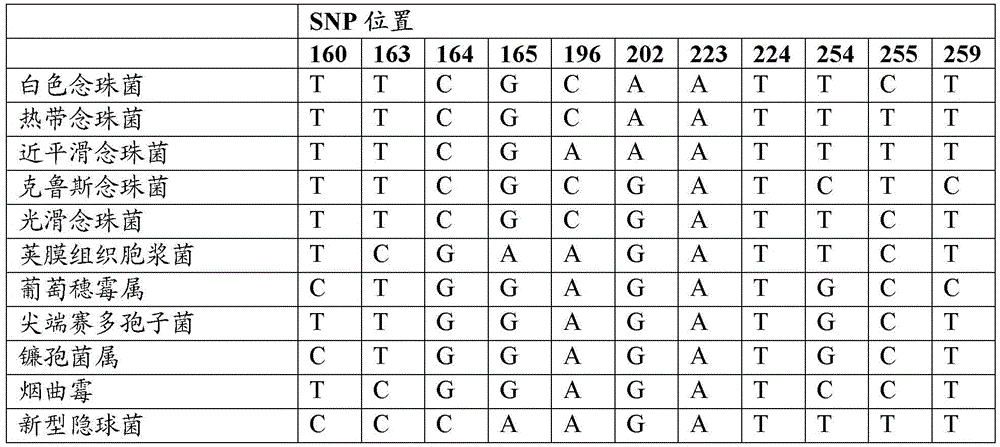
[0392] Identification of 16S rRNA SNPs that distinguish between Gram-negative and Gram-positive prokaryotes
[0393] Representative genes encoding 16S rRNA molecules were downloaded from GenBank and aligned using CLUSTALW to determine conserved sequence regions. Variable sequences as determined by the CLUSTALW alignment were removed, and the sequences were re-aligned using CLUSTALW and checked for any additional variable regions. This process is repeated several times. Subsequently, a final conserved mega-alignment of all genes encoding 16S rRNA was generated, consisting of a conserved sequence region of approximately 702 base pairs. The gene encoding an exemplary 16S rRNA from E. coli (Genbank Accession No. NR_074891) is listed in SEQ ID NO: 1, and a 702 base pair conserved region extends from nucleotides 254-955 of SEQ ID NO: 1.
[0394] The aligned sequences were analyzed and it was determined that two SNPs were sufficient to distinguish most Gram-positive and Gram-negati...
Embodiment 2
[0398] In silico discrimination of Gram-negative and Gram-positive prokaryotes using SNP396 and SNP398
[0399] In silico analysis was used to evaluate which prokaryotes could utilize only SNP396 and 398 and be classified on the basis of their Gram status. Twelve (12) base pair probes (GC(A / C / G / T)A(A / C / G / T)G(CC / TA)GCGT; SEQ ID NO: 2) were used in BLAST searches To identify the region of prokaryotic 16SrRNA spanning positions 396 and 398 (numbering corresponding to the Escherichia coli 16SrRNA listed in SEQ ID NO: 1), and analyze the results to determine which species can be correctly classified as Gram-positive or Gram-positive based on these SNPs. Gram negative.
[0400] Table 31 lists the most common mammalian pathogens that were typed as Gram-negative or Gram-positive based only on SNP positions 396 and 398. Most pathogens can be typed as Gram-negative or Gram-positive based on these two SNPs, except for some Gram-negative bacteria that have A and C or G and C at SNP posi...
Embodiment 3
[0426] In silico Discrimination of Gram-negative and Gram-positive sepsis-associated bacteria using SNPs
[0427] As shown in Example 2 (Table 31), there are several instances where the Gram status of a mammalian (eg, human) pathogen cannot be determined using the SNP at positions 396 and 398. These pathogens include the following Gram-negative genera: Helicobacter, Veillonella, some Bacteroides, Campylobacter, and Chlamydia tropophilia. Although the most common mammalian (eg, human) sepsis-associated bacteria (eg, S. aureus, S. epidermidis, S. hemolyticus, S. hominis, S. saprophyticus, E. faecalis, E. faecium, Clostridium perfringens, Streptococcus viridans group (Streptococcus angina, Streptococcus constellation, Streptococcus intermedius, Streptococcus mitis, Streptococcus mutans, Streptococcus sanguis, Streptococcus aureus, and Streptococcus oralis), Streptococcus pneumoniae , Streptococcus agalactiae, Streptococcus bovis, Streptococcus sanguis, Streptococcus dysgalactiae...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com