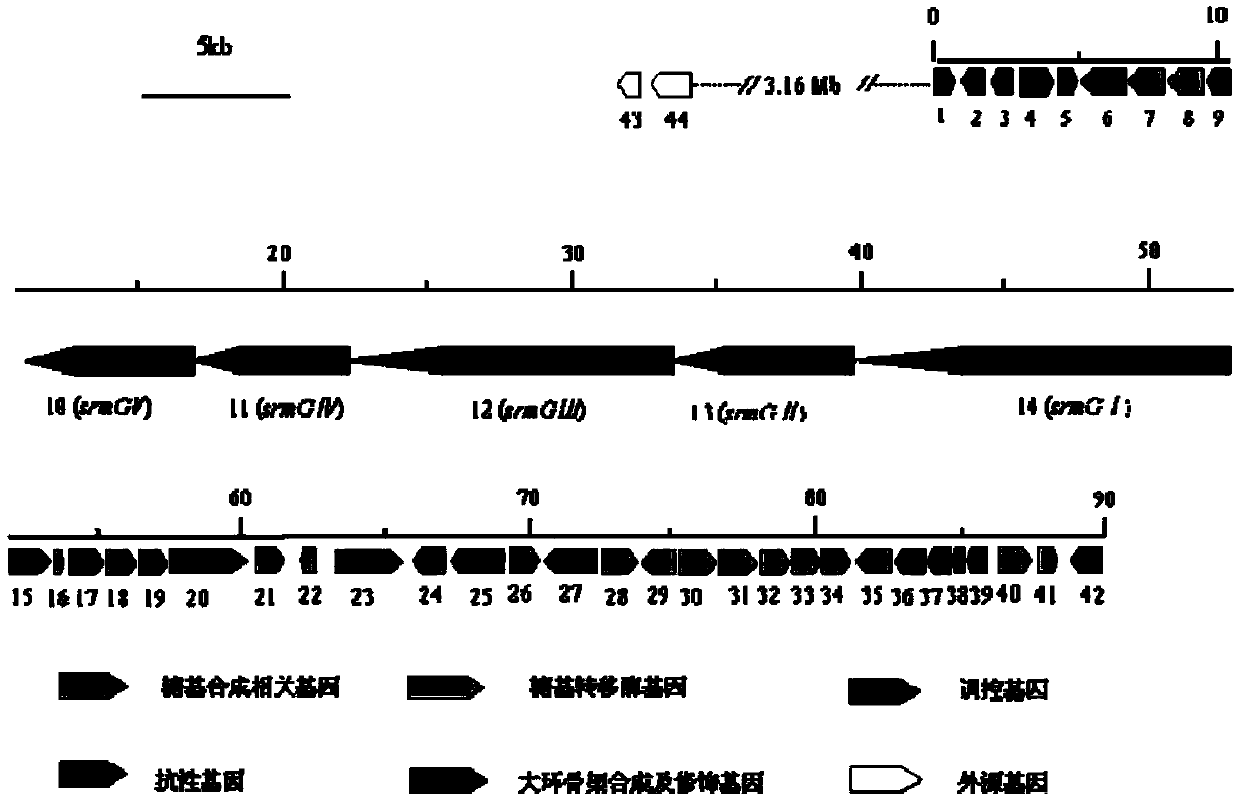
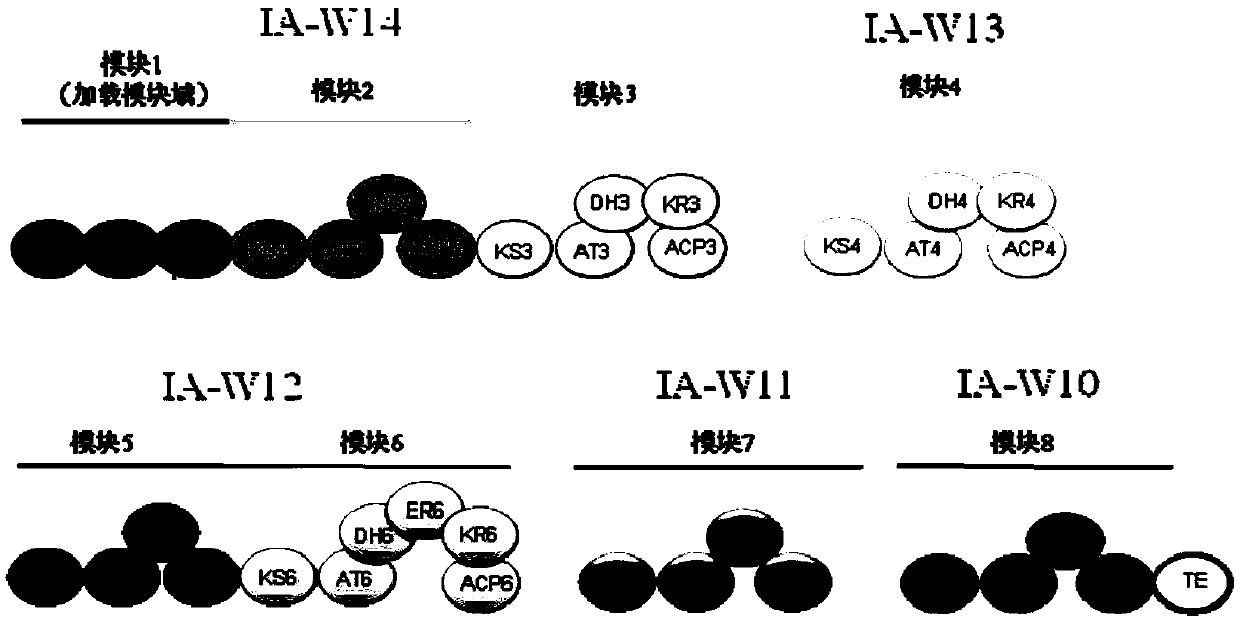
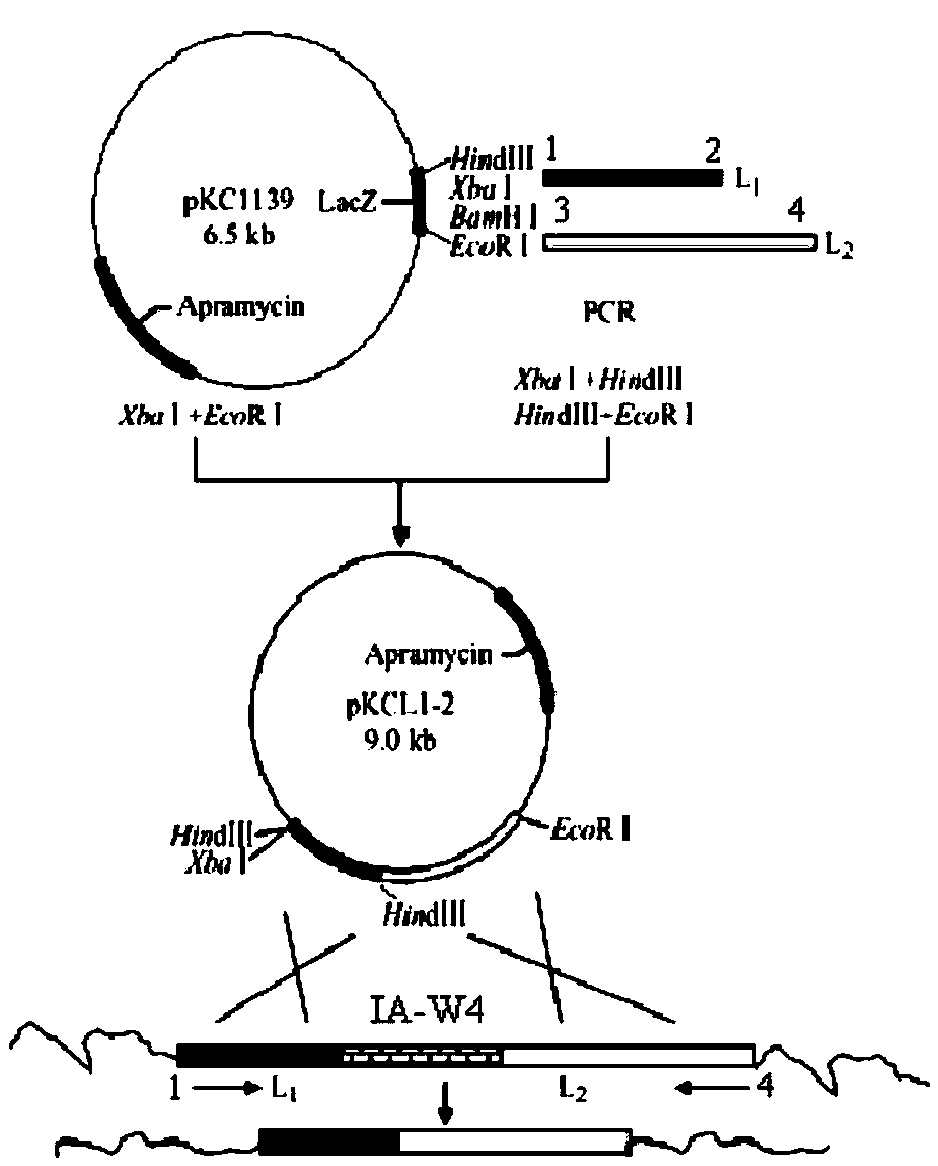
Bitespiramycin biosynthetic gene cluster
A technology of biosynthesis and carrimycin, applied in the field of microbial genetic resources and genetic engineering, can solve the problem of no complete cross-resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] 《Example 1》Crimycin producing bacteria (S.spiramyceticus) total DNA extraction
[0115] R 2 YE medium formula (g / 100ml):
[0116]
[0117] Add 0.2ml of trace element solution, dilute to 100ml distilled water, pH6.5
[0118] Trace element solution (g / 100ml):
[0119]
[0120] 15 pounds of sterilization, 121 ℃, 20min
[0121] S.spiramyceticus strains were inoculated in 25mlR 2 YE medium, cultivated on a shaker at 28°C for 48 hours, and transferred to 100ml R 2 YE culture medium, cultured on a shaker at 28°C for 24h, centrifuged at 5000rpm for 10-15min, collected bacteria (about 10g of bacteria), and mainly operated according to the product instructions of UPTECHTM lifescience company. Add 50ml of 25mM EDTA solution to shake and wash, centrifuge, discard supernatant; use 25ml lysozyme solution (10mg / ml, prepare with Tris-HCl, 2mM EDTA, 1.2% TritonX-100 of 10mM pH8.0, add 0.5ml100mg / ml mlRNase) suspended mycelium, cultured at 37°C for about 1-2h, and cultured ce...
Embodiment 2
[0122] 《Example 2》Verify the function of Seq.1 gene information by gene blocking
[0123] IA-W1 and IA-W42 as well as IA-W4, 17, 21, 23 and 27 genes at both ends of the gene cluster were selected for blocking to obtain mutant strains, and experiments proved that the ability of these blocking strains to produce corimycin occurred changes, or no longer produces corimycin. Thus it is suggested that the obtained gene cluster information is necessary for the production of carrimycin. Primers were designed according to the above coding genes and their upstream and downstream sequences, and inserted into appropriate restriction sites. The primer sequences are shown in Table 2.
[0124] Table 2 Primer sequences designed and used in gene blocking experiments
[0125]
[0126]
[0127] The corresponding homologous gene fragments were respectively obtained by PCR amplification, and the corresponding restriction sites were used, and the selection marker resistance gene (Apramycin-...
Embodiment 3
[0131] "Example 3" Gene transfer of corimycin-producing bacteria and screening of blocking strains
[0132] 3.1 Preparation of protoplasts: inoculate the slant spores of fresh carrimycin-producing bacteria in R 2 In YE liquid medium, 28°C, 220rpm shaking flask cultured for 48h, the culture solution was transferred to fresh R with 0.5% glycine supplemented with 10% transfer amount. 2 In YE liquid medium, shake culture at 28°C for 20h. Take 10ml of bacterial liquid in a centrifuge tube, centrifuge at 3000rpm to collect mycelia, and use P-buffer for precipitation
[0133]
[0134] Sterilize at pH7.615 lbs, 121°C, 30min.
[0135] After washing twice, suspend with an appropriate amount of P-buffer, add the P-buffer solution of lysozyme (final concentration is 2mg / ml), mix well, keep warm in a 37°C water bath for 30-45min, and shake once every 10-15min. Use a 10×40 phase-contrast microscope to observe the formation of protoplasts. When the microscopic examination shows that mo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com